

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_002840 | SP | 0.007208 | 0.992779 | 0.000014 | CS pos: 14-15. INA-IS. Pr: 0.7711 |
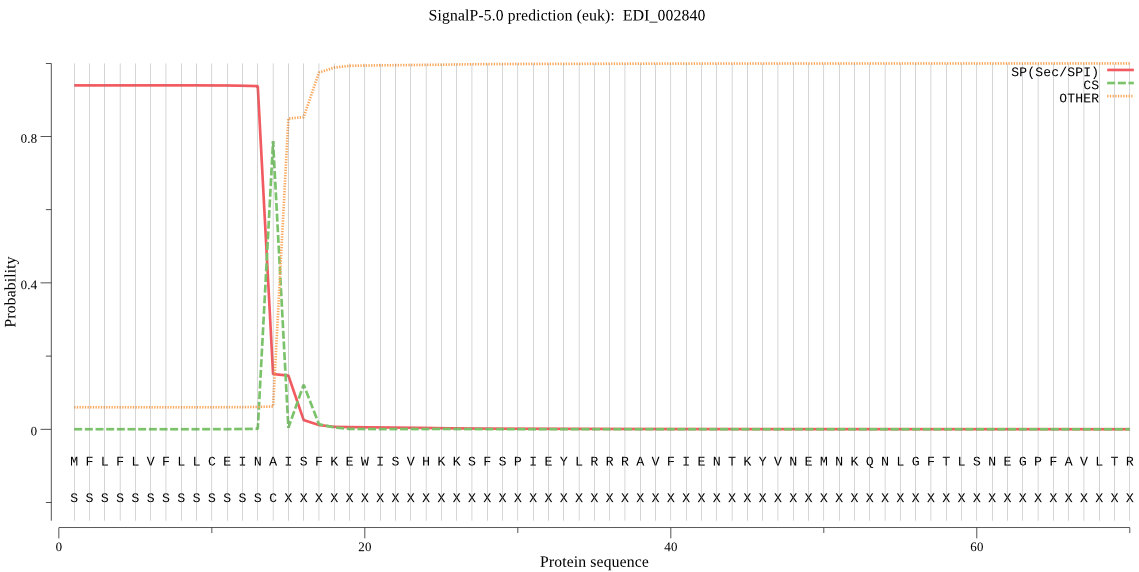
MFLFLVFLLCEINAISFKEWISVHKKSFSPIEYLRRRAVFIENTKYVNEMNKQNLGFTLS NEGPFAVLTREESIAVTQGFRMDKSDLEQHKTLKREMTEAIDYRNIQGKNYMTPVKDQEN CGSCYAFSSVALMETAVLLAYDDLSPNTYALSTAEIVSCCYDPNGCRGCEGGSIGSALKY AQDNGMQTESSFPYKPFEQRCLQGEKVMKVKKYTHSDTKGDDEKVRSEILSYGPVGSSMD ASRSSFLLYHGGIYNDKKCRSDKPTIAVVIIGYGIDKSNNKYFIVRNNWGQYWGEQGYFR ISSDNNLCGLSNDIYYIESIERLK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| EDI_002840 | 29 S | KKSFSPIEY | 0.993 | unsp | EDI_002840 | 261 S | KKCRSDKPT | 0.993 | unsp |