

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_012640 | SP | 0.022577 | 0.976738 | 0.000686 | CS pos: 14-15. SFS-QE. Pr: 0.6074 |
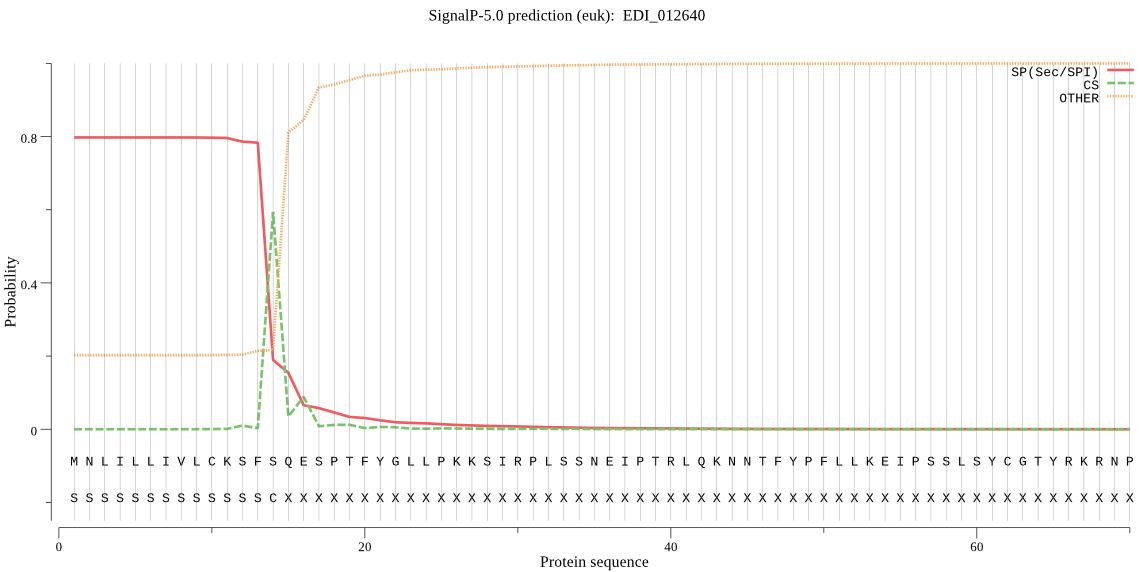
MNLILLIVLCKSFSQESPTFYGLLPKKSIRPLSSNEIPTRLQKNNTFYPFLLKEIPSSLS YCGTYRKRNPHKIEDYCIKQTMNQGECGGCYAMALAQVLQAHYTHLTLRHQAFSTQQIID CSNNNGCSGGWPTSVLESLQYFVSEIEYPYRLITINGNSTNNYKKECIRGRRTRIHISDY KEVIGSISFEQIKVLLIEHGPFIGMIYSNDQLRKYSGGILHLNCPVVPTLNHAIIVVGYG EENQEKYIIIRNSWGNSWGEMGYARISINDLCGLDGIKYSDYPTILYLKTQLPKQIYSNY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| EDI_012640 | 33 S | IRPLSSNEI | 0.995 | unsp | EDI_012640 | 267 S | YARISINDL | 0.993 | unsp |