

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_020670 | SP | 0.003151 | 0.996826 | 0.000023 | CS pos: 18-19. SYA-LY. Pr: 0.9534 |
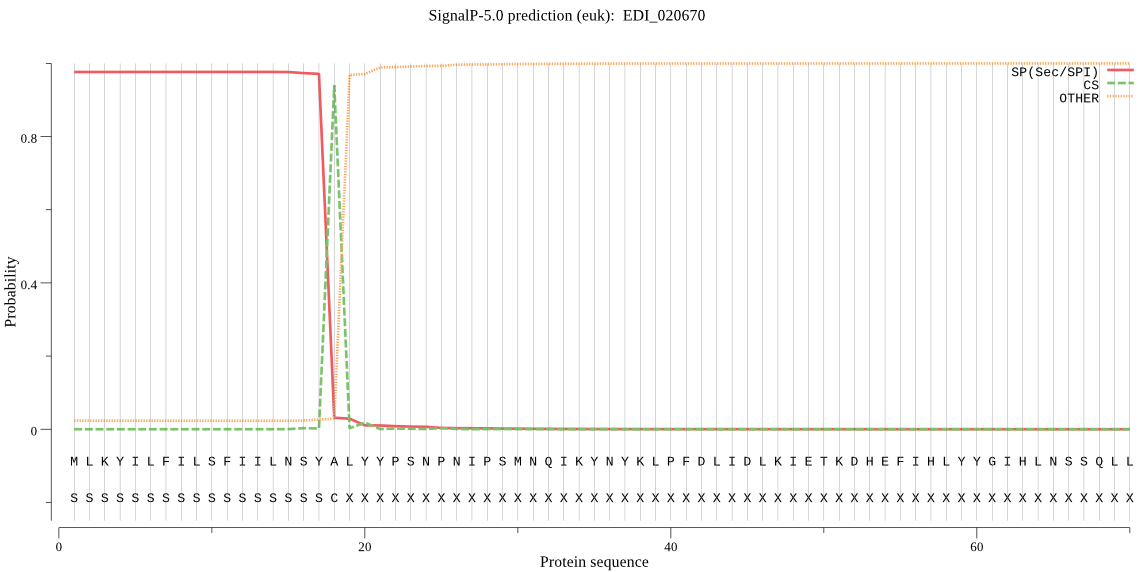
MLKYILFILSFIILNSYALYYPSNPNIPSMNQIKYNYKLPFDLIDLKIETKDHEFIHLYY GIHLNSSQLLLLFQSNAGCFLDRLFLLKSFYSKFNISVGILSYRGYGNSTGKPSEQGFIE DALASLSYLNKEGIPIQNIIIIGRSIGVGVALSITQILPIKKLILENGFTSLIEFLPSLQ NNEVMIRDPWLNEQKIETINKNTSILFLLSEEDEIVPTWMTRKMEKKARDLGIQTKLVSF PGAKHMQLPYYDNYYRVIKEFLNEQKQRGLSK
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EDI_020670 | 210 S | LFLLSEEDE | 0.995 | unsp |