

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
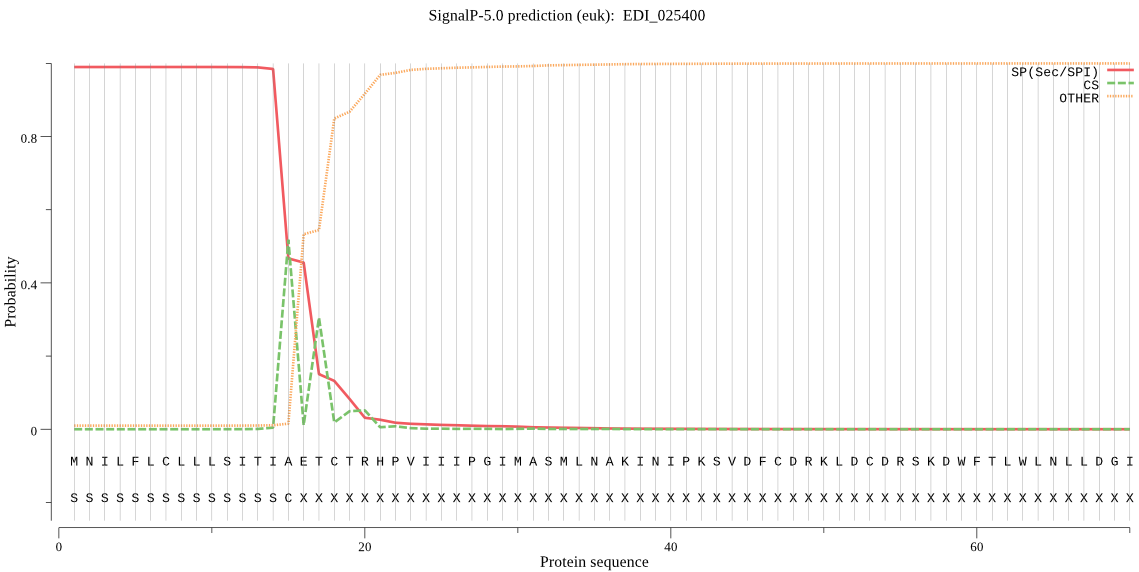
| EDI_025400 | SP | 0.000211 | 0.999785 | 0.000004 | CS pos: 15-16. TIA-ET. Pr: 0.6452 |
MNILFLCLLLSITIAETCTRHPVIIIPGIMASMLNAKINIPKSVDFCDRKLDCDRSKDWF TLWLNLLDGIPYVNDCYIAYLTCHYNSTSGRMENVEGVQIEPPRFGSTYAVDTISPSFPL KTFSRAFHEIIKGLEKIGYKDEFDLFSAPYDWRYYHHDEYYEKVKELIIKAYENTGNKVV LVSHSMGGLTTYILLDKLGKEFCDKYIHRWVAMSTPFIGTTIANDIVLSGYNMGYPVSKE LIKKASRTFETVAMMGPIGEYWDQNEVLVELANGKKYYPKDQVELFSQLEEMKPFAKEAV ENSFAPYLKKYNSQVPHGVEMHCGITSGIETAHKITFKGDTFDSKSTIEFVDGDQEVTLN SLEXXTLNSLEYCKKMTENFTNLGKYTHQGMLLKKEVIEYVKNLACD
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EDI_025400 | 346 S | FDSKSTIEF | 0.994 | unsp |