

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_028230 | OTHER | 0.985552 | 0.000863 | 0.013585 |
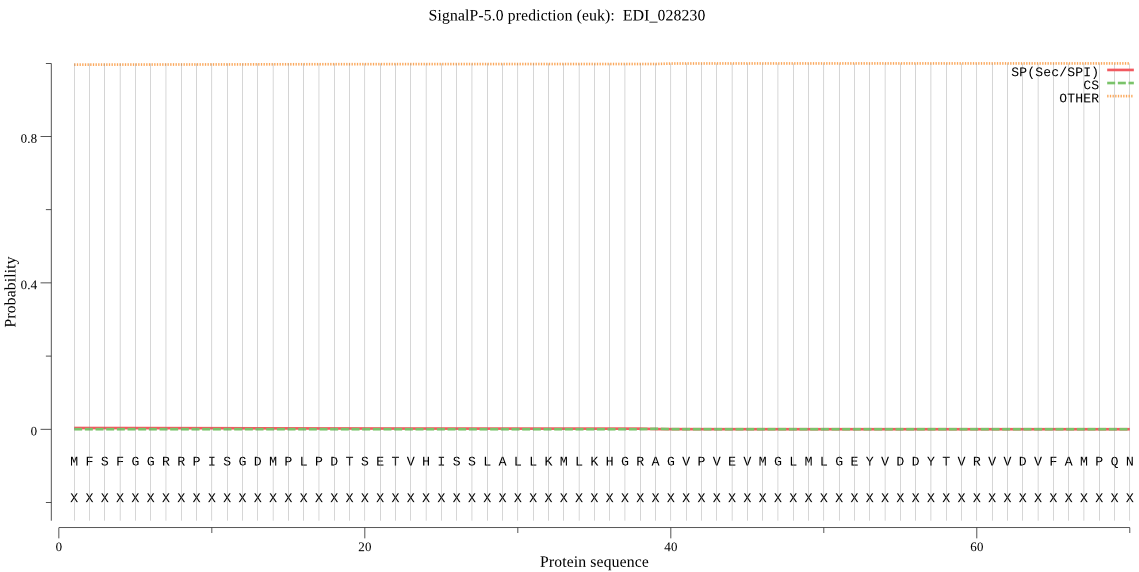
MFSFGGRRPISGDMPLPDTSETVHISSLALLKMLKHGRAGVPVEVMGLMLGEYVDDYTVR VVDVFAMPQNGTGVSVEAVDEVYQTTMIEMLRQTGRKESIVGWYHSHPGFGCWLSSIDIS TQQSFEKLNERCVAVVVDPIQSVKGKVVIDAFRTIQNQFNLGVEPRQVTSNQGHLTKPTS QAKVRGLGKQYYSMPIEFSKNEIDERMLLNLQKKKWTDSLEVESSQRKDSNIQTIDKLIG LMKQYNQNIQDEALDPKLRDLQQVGKIDPKKHLQRIADQALTENAVDTLGVFMNTVVF
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EDI_028230 | 230 S | QRKDSNIQT | 0.993 | unsp |