

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_029570 | SP | 0.012801 | 0.987190 | 0.000009 | CS pos: 15-16. SEG-SS. Pr: 0.6109 |
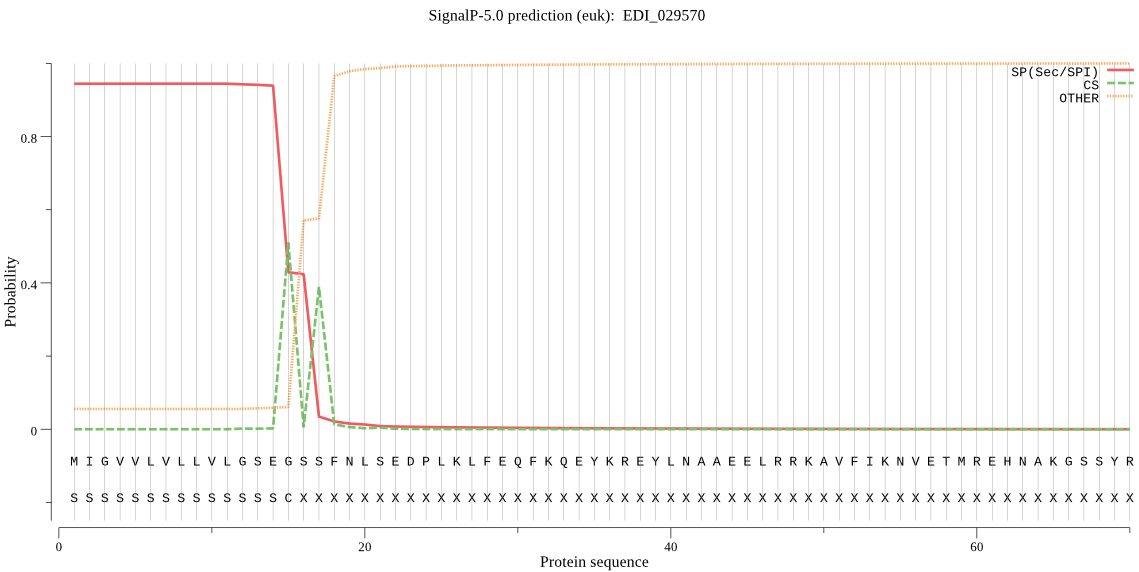
MIGVVLVLLVLGSEGSSFNLSEDPLKLFEQFKQEYKREYLNAAEELRRKAVFIKNVETMR EHNAKGSSYRMGINKFADMEDSELLAASSRIQNNRRMGAKRIQEMIKMEKVSPTKQTLKQ AVPANYTLCTSAAEYNYCGTNNINQNTCGGCYAVGPAQHLSTIYAYLTKQANNDKPKRKL LSAQQLLDCNTVAWGGCGGGFAEDVLNSINGIYYADDYPFIDADKDCEDPEDCEKFKHEC KSLEVEYPLKFATENKFTHFDNKNWEEIKEIIYKYHGFISAMKVTSSLYRYEGGIYKSES CTGQITDHVITVDGYGECDGHKFLWVRNSWGNDWGIENGHFKIDWNTLCGINGVDVFRNG NSLQNNLYVEMELGDGDIAEYNPEDGAGDDNEDPIGCTIVPSSGDPSNGPDDNKTTGVVM VLFIFLMIFFI
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| EDI_029570 | T | 398 | 0.500 | 0.404 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| EDI_029570 | T | 398 | 0.500 | 0.404 |