

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
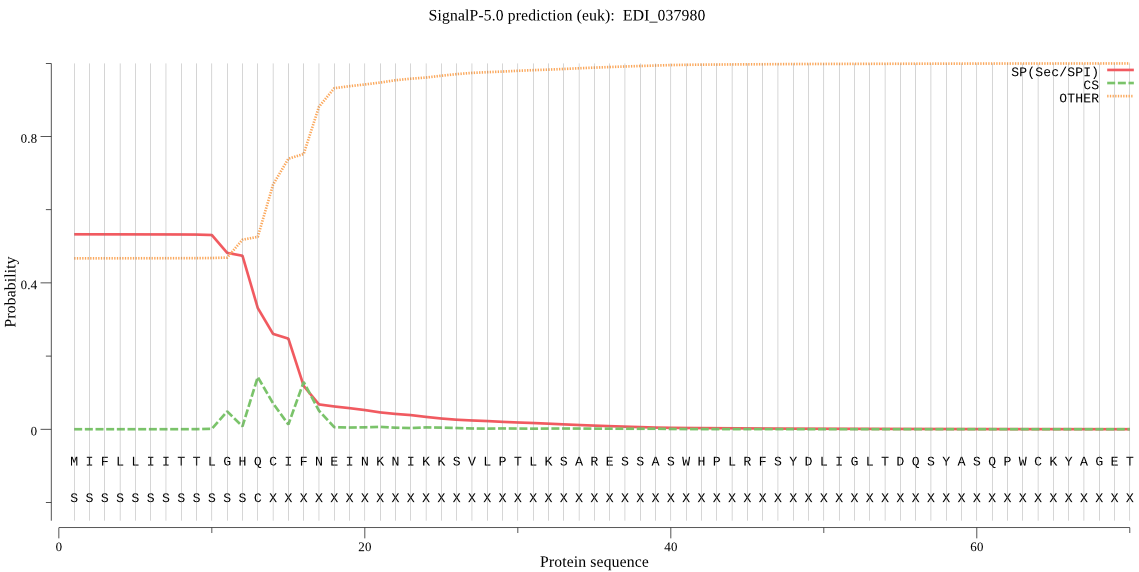
| EDI_037980 | SP | 0.115082 | 0.884219 | 0.000699 | CS pos: 14-15. HQC-IF. Pr: 0.2979 |
MIFLLIITTLGHQCIFNEINKNIKKSVLPTLKSARESSASWHPLRFSYDLIGLTDQSYAS QPWCKYAGETIVLSSSNIYKCSEDDVLTKEKIEYIDRLFQSLNQSISEIFNVHTINYNKV IPGKKQCGEAVFPRAYTIPEESDFHIMVTSHPHTSSSVFAYAAACYYGIDSQVNKRRSLM GYVNIVTSALEPTEERFRDQVGIIFHETMHALGFDGSLGVDKKVGNLNLKVVEDEVVVVR ARKHFGCDNMTYVPLEDGGGPGTANAHWERRIFISEIMNGITSNNPRISEITLAYFEALG VYKPNYEMADALSWGRGVGCSFLEDCSKWPKQNGYYCRTGVRRCTNDRSAIGICDGNSFK ETLNQSYQHYGDPHVGGSDEIADNCIFVSPIETSYCYKKMSLLSFDYYFTLEMLNHGQNF GDTSMCFESSLIKFIPYGINNYHCYSVACMNNSFYKISIDGRYYDCDKSIKVRGYGGKLI CADPKDICGNRVIETWPEIDSVENKTISVGDILFINGRNLDVVQKVYIDYTSCKIINKSK EQLQIEVDYRDPFISFTSFEASLMVKVNESVNSCYDHLIKIKVPYHYIFLNFGDWVMTNY IFIAVIPVWILFFILLFGYYILKVIVIRQAEEVHEAKREINLRLNKFDNKPQKKKQCFCV MF
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDI_037980 | 508 S | NKTISVGDI | 0.992 | unsp | EDI_037980 | 508 S | NKTISVGDI | 0.992 | unsp | EDI_037980 | 508 S | NKTISVGDI | 0.992 | unsp | EDI_037980 | 40 S | ESSASWHPL | 0.992 | unsp | EDI_037980 | 358 S | CDGNSFKET | 0.995 | unsp |