

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
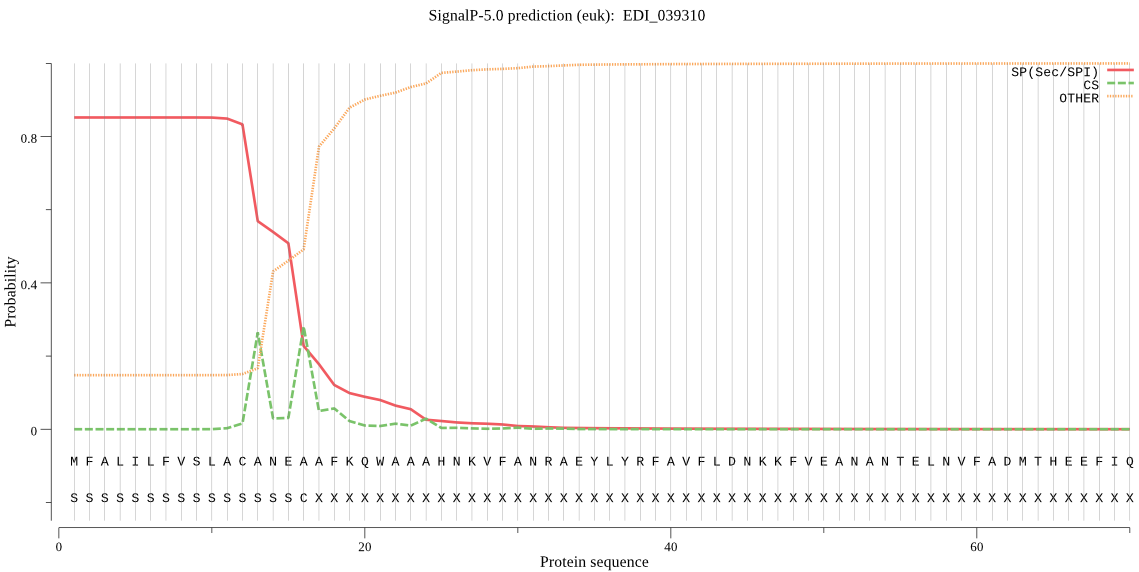
| EDI_039310 | SP | 0.001771 | 0.998191 | 0.000038 | CS pos: 16-17. NEA-AF. Pr: 0.3995 |
MFALILFVSLACANEAAFKQWAAAHNKVFANRAEYLYRFAVFLDNKKFVEANANTELNVF ADMTHEEFIQTHLGMTYEIPETTPSVKAAIKAAPESVDWRSIMNPAKDQGQCGSCWTFCT TAVLEGRVNKDLGKLYSFSEQQLVDCDSSDNGCEGGHPSNSLKFIQENNGLGLETDYPYK AVAGTCKKVKNVATVTGSKRVTDGSETGLQTIIAENGPVAVGMDASRPSFQLYKKGTIYS DAKCRSRMMNHCVTAVGYGSNSNGKYWIIRNSWGTAWGDAGYFLLARDSNNMCGIGRDSN YPTGVKLL
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| EDI_039310 | T | 82 | 0.508 | 0.234 | EDI_039310 | T | 83 | 0.503 | 0.175 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| EDI_039310 | T | 82 | 0.508 | 0.234 | EDI_039310 | T | 83 | 0.503 | 0.175 |