

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
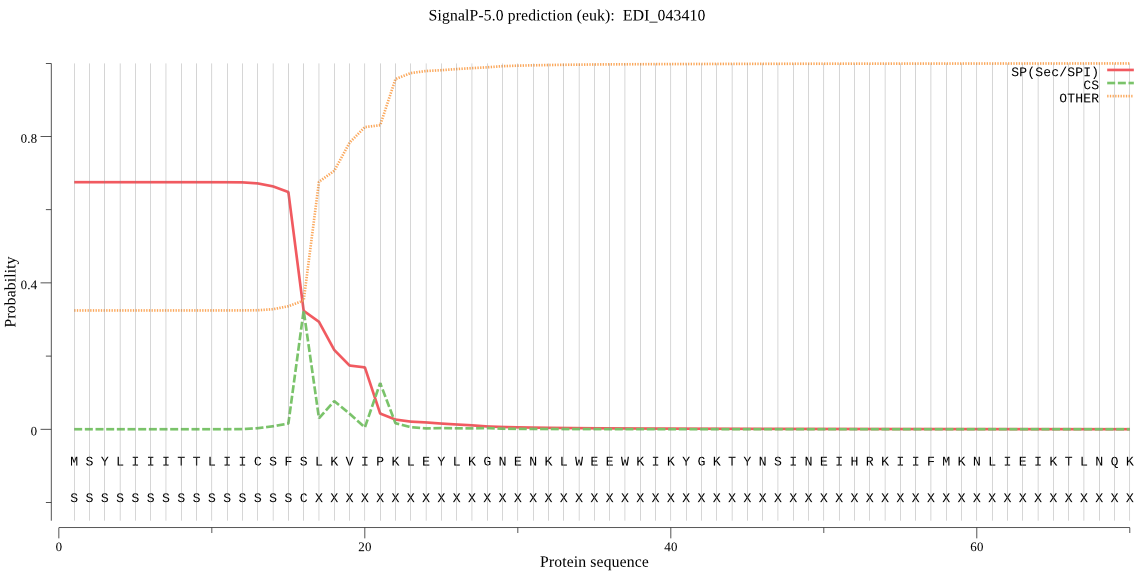
| EDI_043410 | SP | 0.029494 | 0.970312 | 0.000194 | CS pos: 21-22. VIP-KL. Pr: 0.3920 |
MSYLIIITTLIICSFSLKVIPKLEYLKGNENKLWEEWKIKYGKTYNSINEIHRKIIFMKN LIEIKTLNQKREKDIDAYFDLNQWSDLSNQEFEDLMLMKKPKRNNVKELHNKINITIPYP KGPVPINYSACNQKMLFGKLNPGEIDFCNGTEFDQQSCGSCYCVSNALALQLKWANLTYL RDGKPQYTMFSPQQFLDCEVGGYHCAGGYTDGPLDFSHYVSTIDDYPYYSGREPSKRKTC VKGKRTPIKFSYTIFDSAEDINIIKPIVHHYGGFVSCVYPKYWTAYRGGILRGLNCEKGV ITTHVVGVVGYGIEDGIEYVVVRNSWGKNWGLGGYIKLGADSLCGIGGNDGGDVPVSVVL HTDFSDVEYGPYGEFRNNTDVPQRLYNESADTNESSTNIESNDEASSESQSLNNHQSEES LVVEESSSVEPNIRPKNENRLRSIMIYILYLLVGISVITMVVAVIILITQ
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDI_043410 | 235 S | GREPSKRKT | 0.995 | unsp | EDI_043410 | 235 S | GREPSKRKT | 0.995 | unsp | EDI_043410 | 235 S | GREPSKRKT | 0.995 | unsp | EDI_043410 | 406 S | NDEASSESQ | 0.991 | unsp | EDI_043410 | 221 S | SHYVSTIDD | 0.994 | unsp | EDI_043410 | 230 S | YPYYSGREP | 0.993 | unsp |