

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_057770 | SP | 0.003192 | 0.996807 | 0.000001 | CS pos: 14-15. CKA-EF. Pr: 0.9423 |
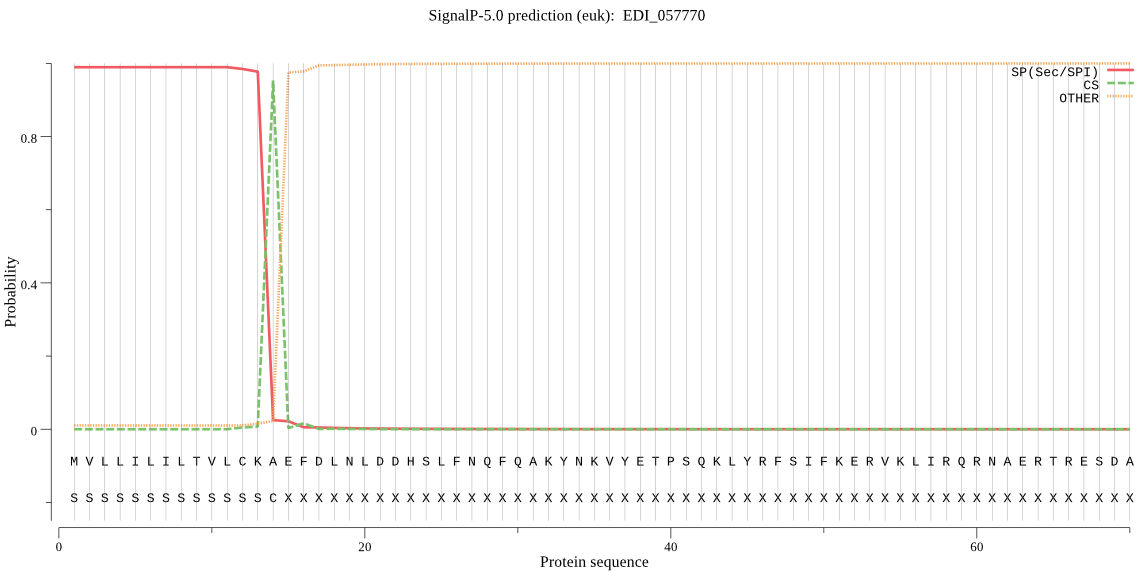
MVLLILILTVLCKAEFDLNLDDHSLFNQFQAKYNKVYETPSQKLYRFSIFKERVKLIRQR NAERTRESDAIFVINALTDLYPAEYGVVPPIPIDISKLKGYDDHYNRDYNEPPPLPEGDI LPVYHTYCGKYVVNNTKRDAVDLCGKQFNQGSCGSCYAASTANLGQYLYANISYYYNDKD INKTVKPLFTPQRWIDKAIPGGTNLQISRRCCGGNPMNILDAEPSYSLESDYPYIDGSGS TSTSCSPRGDQNPNVKIQMRNDHYHVFAVTGTHEEKVLTLKKILHHYGPISTAIYSNADV LFANAGSGIYTFPSTCTDTTSTDHQVILVGYGKEDGREFFIMRNSWYDGWGDGDNYLKIS TDVLCGIGQKIGDYIIPLNYIVFAGNCKLDKNCNSCDEKTLVCSSCKENTTLDARGMCVG EWEDPYANVDDGSVSILLNMSILLILLFFMI
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDI_057770 | 426 Y | WEDPYANVD | 0.992 | unsp | EDI_057770 | 426 Y | WEDPYANVD | 0.992 | unsp | EDI_057770 | 426 Y | WEDPYANVD | 0.992 | unsp | EDI_057770 | 345 S | IMRNSWYDG | 0.996 | unsp | EDI_057770 | 405 S | LVCSSCKEN | 0.992 | unsp |