

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
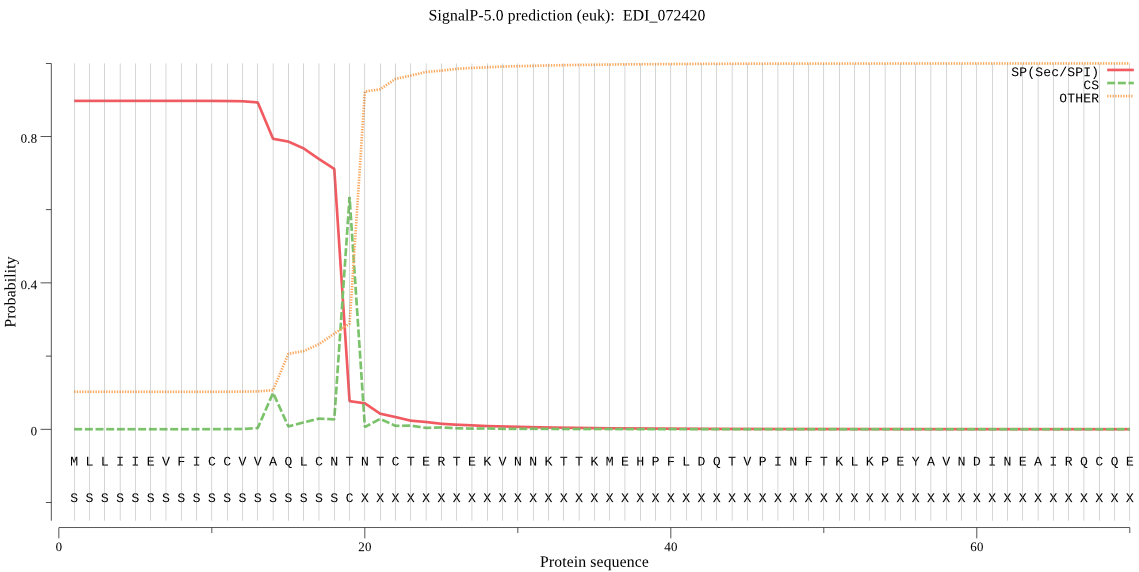
| EDI_072420 | SP | 0.031854 | 0.964275 | 0.003870 | CS pos: 19-20. CNT-NT. Pr: 0.7282 |
MLLIIEVFICCVVAQLCNTNTCTERTEKVNNKTTKMEHPFLDQTVPINFTKLKPEYAVND INEAIRQCQEQINQIKNIKIEEASFENVVRAFDNATIILDDSMLKLEVLVSCRSDIDEYR KAHEEVLPKVTEFRATLHSDEIIFSLVKKVYENKENLDYDQKRLVEETYEDFIEGGANLN KEDKTKLVELKKEITQLTKDFSNNLVDAVDEYTMNVEDKNKLEGIPENIIDMYKKEAEDR KKEGYIITLQVPSYFPAIKYCKNEEIRKELFIQYNKLCSEGKFDNTKLIQEIIEKRQKYA ELLGYHDFSDLVTKRRMVKNGTNAIEFINQLHDKVQSFFYKEEEELKEFKKEKCGNNQLE PWDVNYYIELMKQEKYNFNEEEFREYFTEESVFGGLFEIYKRLYGVSFKLNKEIKGWHDD VKYYEVIEKDGSIIGGVYFDLYPRKGKRSGAWEEMLRVSYENTNGTFIKPLGIVCTNITP PINGITRLNHDEVETIFHESGHLMHTLFTKQRYASLAGTNVAWDFVELPSQIMENWTFER EALDLFSQFGNVKPIPKELFDKMWKAKNYMSAYYTMRQLSFGKIDLEIHRNYLTSGLSLD DFIYQKEKDYKYHFNTKTLSIIRVFQHLFSHFTGYACGYYSYKWAESIDADAFEYFKENG IFNQSIANKFRTNILSMGNSKPADELYRNFRGRNPDFTALLRRAGLLEKK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| EDI_072420 | 459 S | MLRVSYENT | 0.995 | unsp | EDI_072420 | 580 S | MRQLSFGKI | 0.991 | unsp |