

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_079160 | SP | 0.232469 | 0.674834 | 0.092697 | CS pos: 12-13. SLG-FR. Pr: 0.3909 |
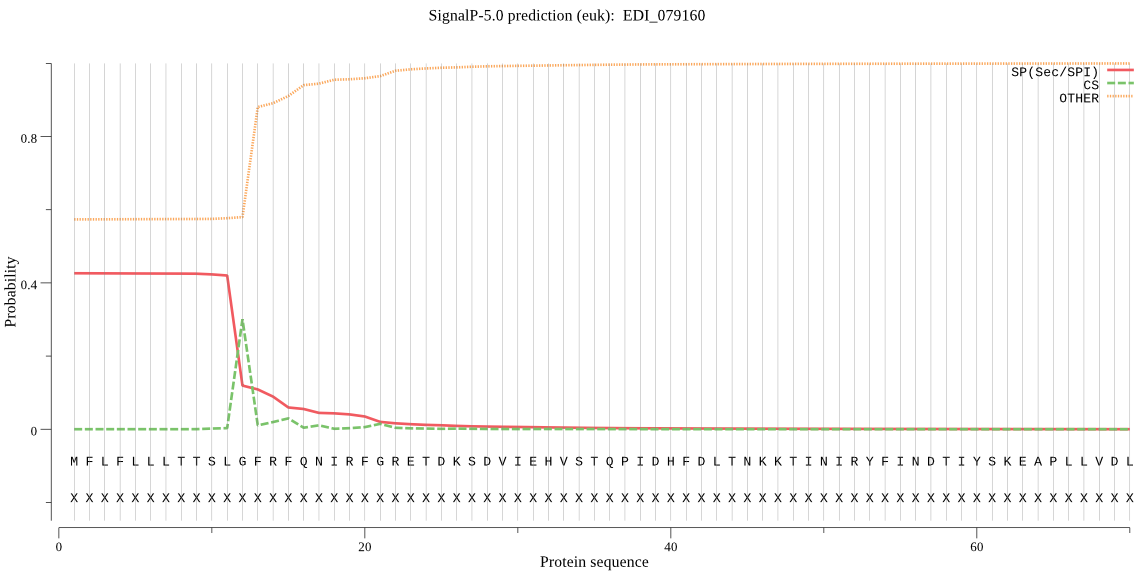
MFLFLLLTTSLGFRFQNIRFGRETDKSDVIEHVSTQPIDHFDLTNKKTINIRYFINDTIY SKEAPLLVDLGGEGPQKAAAVGGRFVINKYAEKYNSLMLAIEHRFYGKSVPEGGLSQENL GYLSAAQALEDYIMIINQIKKEYQVTGPVIVFGGSYSGNLAAWIRQKYPNVVYAAVASSA PVYATSTFYEFLDVIYNDMGEKCGNAWKEATESIEELFKTDSGKAQLKSDFNACTDINGE DDLTILIQQIQATMINYPQYNGSYSLTIEGVCSILTTEGKTAYENMVELMNHAFNEFGFK CAPSSYADMLTDMANTKTDEEGNRLASTRSWAWQICSEYSYFQPVNESLPFSKRLNNEFY YLLCKDIFNVDKQRLDRRVHHTNLMYGGYQPKATNVAYTSGSTDPWSPLAKHETLPSDIN CYASHIKGTAHCADLYAEKDTDPEQLKQQRIETAQFIDELISRYHN
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EDI_079160 | 213 S | EATESIEEL | 0.995 | unsp |