

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
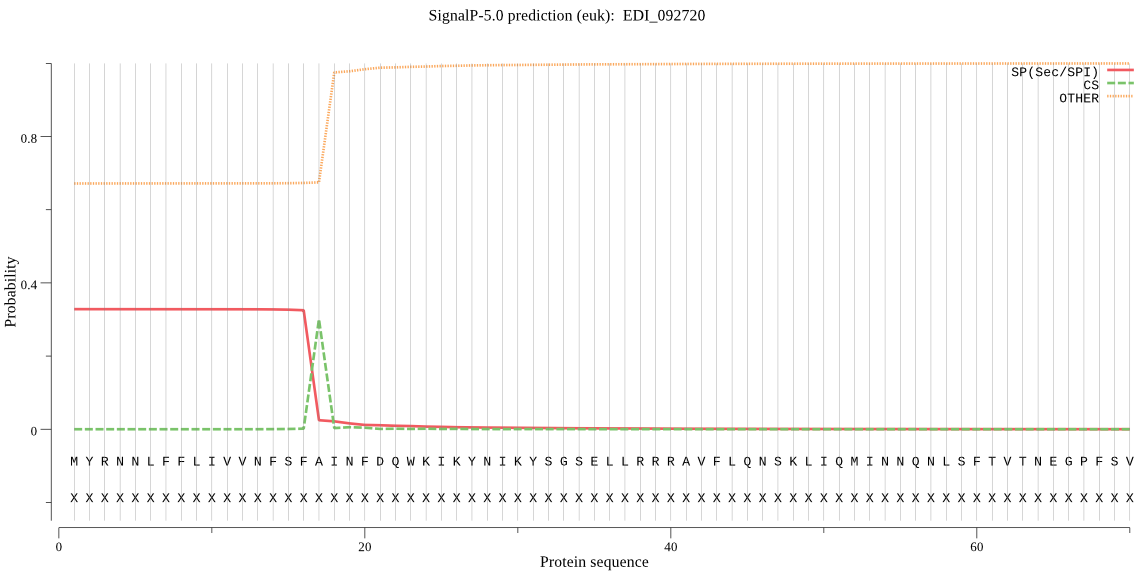
| EDI_092720 | SP | 0.066978 | 0.930707 | 0.002316 | CS pos: 17-18. SFA-IN. Pr: 0.7393 |
MYRNNLFFLIVVNFSFAINFDQWKIKYNIKYSGSELLRRRAVFLQNSKLIQMINNQNLSF TVTNEGPFSVLTNEEYKMLHHRIDIEKEIKQLKSHKMNFLKNIDNKEVLDSIDWRSEGKV TPVKNQRKCASCYAFGSVATIESLIMQETHIKEIDLSEQQIVDCSQGEYSNWGCTCGNVG NSFNYIRDHGVLLERDYPYTGKANNCSIDGKKPVIKIKDYSFVFPQTEENLKIAVYHQPV AVSIDSSQPSFQFYEGGIYDEPNCKWVDHIVTVVGYGTTEEHQDFWIVKNSYGNEWGLNG YIYMSRNKNNQCGIATIAVYPKGLIIL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EDI_092720 | 94 S | KQLKSHKMN | 0.994 | unsp |