

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
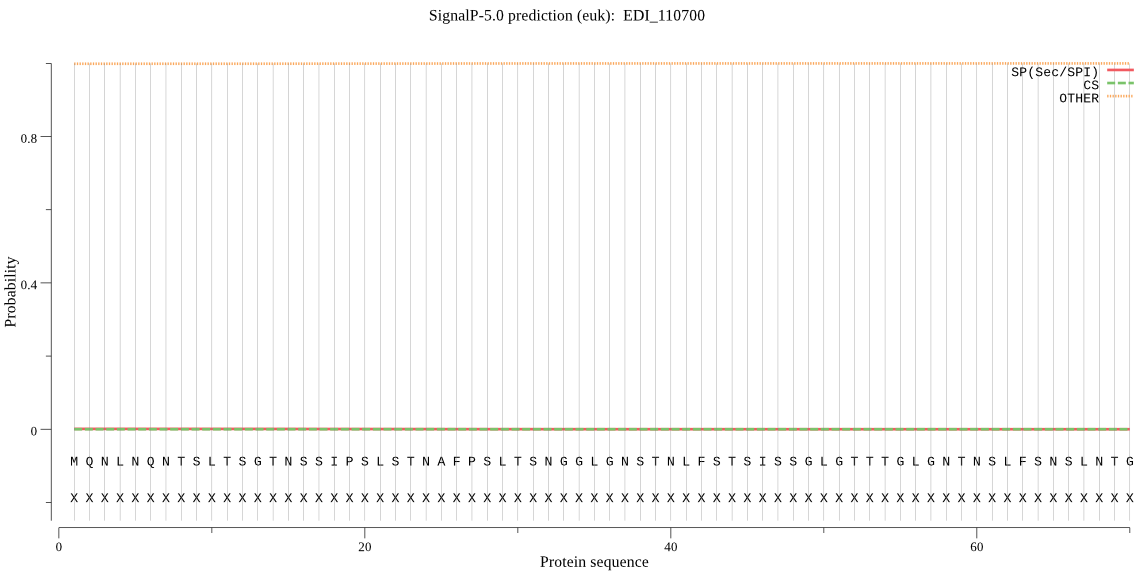
| EDI_110700 | OTHER | 0.999539 | 0.000084 | 0.000377 |
MQNLNQNTSLTSGTNSSIPSLSTNAFPSLTSNGGLGNSTNLFSTSISSGLGTTTGLGNTN SLFSNSLNTGFGTNTTTGLGTTNSLFSNTFNTGLGNSTTGNNNPILLNPLSISSLPIGTN ASKDKQSKEKSWVYHDQTSMNQPDFVRPEEMMFNTSCCIIDPKVIDKILFDAEREIEALQ PNISYDIKPIGLKQGYVCNPSMQTLKAMSIEQLKNVSSFEVSRPGYGKVMWSNVDLTNVN LDISIIIERGYCDVYPDGIQKPRFGEKLNKEATIVLENVVDTLEEFRKLLPRLSKKVDIK SYEENTKTVTFVVPHFTRYSVIDEDNNTQENEEEVIDYEKHQTISNNNPEWKFPVVVLHK IEFSPFEIDI
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| EDI_110700 | T | 23 | 0.535 | 0.108 | EDI_110700 | T | 8 | 0.502 | 0.026 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| EDI_110700 | T | 23 | 0.535 | 0.108 | EDI_110700 | T | 8 | 0.502 | 0.026 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDI_110700 | 320 S | FTRYSVIDE | 0.994 | unsp | EDI_110700 | 320 S | FTRYSVIDE | 0.994 | unsp | EDI_110700 | 320 S | FTRYSVIDE | 0.994 | unsp | EDI_110700 | 209 S | LKAMSIEQL | 0.995 | unsp | EDI_110700 | 301 S | VDIKSYEEN | 0.997 | unsp |