

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_113060 | SP | 0.057605 | 0.941649 | 0.000746 | CS pos: 13-14. ANA-IT. Pr: 0.4618 |
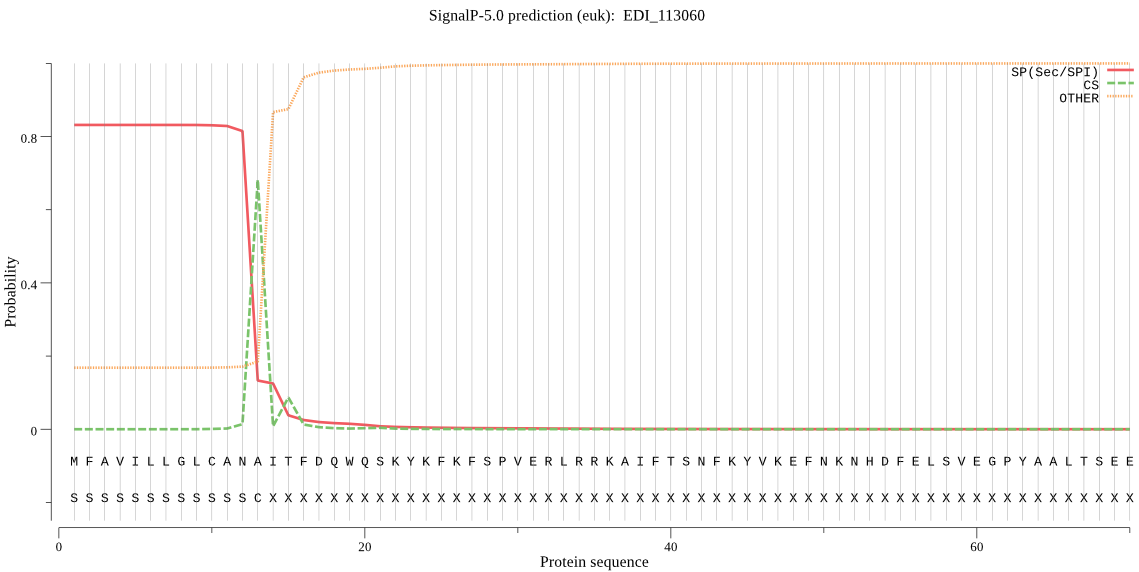
MFAVILLGLCANAITFDQWQSKYKFKFSPVERLRRKAIFTSNFKYVKEFNKNHDFELSVE GPYAALTSEEYQALLNKQIIGNENKNIEVINLKSNKQVPASVDWRAEGKVTPIKDQGSCG SCFTFGAVAAIESRLLISGTTGYTAQTLIFLNNKFLIVLFLLMDDNPYTGKDGKCLYDKT KVKVSIEGLRTADISDTALTAAIADAPVGVCIDASQTSFQLYKSGVYDEPKCKKIMNHCV AAVGYGSQDGKDYYIVKNSWGTTWGMDGYILMSRNKNNQCGICTGISFPVGVKLI
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EDI_113060 | 185 S | KVKVSIEGL | 0.996 | unsp |