

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
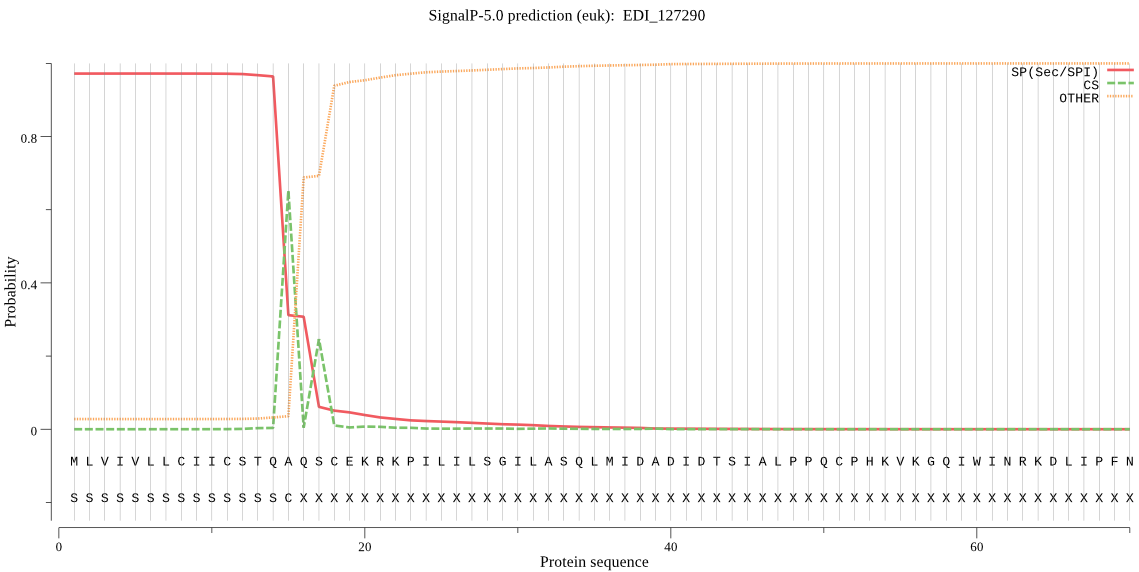
| EDI_127290 | SP | 0.002607 | 0.997248 | 0.000145 | CS pos: 15-16. TQA-QS. Pr: 0.9087 |
MLVIVLLCIICSTQAQSCEKRKPILILSGILASQLMIDADIDTSIALPPQCPHKVKGQIW INRKDLIPFNNSACFVEYMKTYWNSSTRKMENIPGANIYYPDFPSTKGIFALAPDNQKLL QSKTKVFAAMIRDLKAAGWKDGIDLVSPGYDWRYADRSNNNWTEKTTQLIQQLVHDNGHK VVIVTHSFGGIAVLDLISSMSKEFCDQYIDKIITLNAPFIGSTKTLRTFLTGEDLGLKLD PLLLRPLARSWESDYQLMPNQRYWKNDNIVQVGNKKYSANNINAIIDLVEEVKEFGNIIY NSSINRHPLEYVPNNVTLHCLYSHGIETIVGIKYDSLDHDFQDVSYVYGDGDGVVDLQSL EWCKLPGFAKVVKDLGKGEHGTVISNTEVFGYIKNEACARDSNTKM
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EDI_127290 | 250 S | PLARSWESD | 0.994 | unsp |