

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_156850 | SP | 0.014792 | 0.984548 | 0.000660 | CS pos: 20-21. SWA-AK. Pr: 0.6209 |
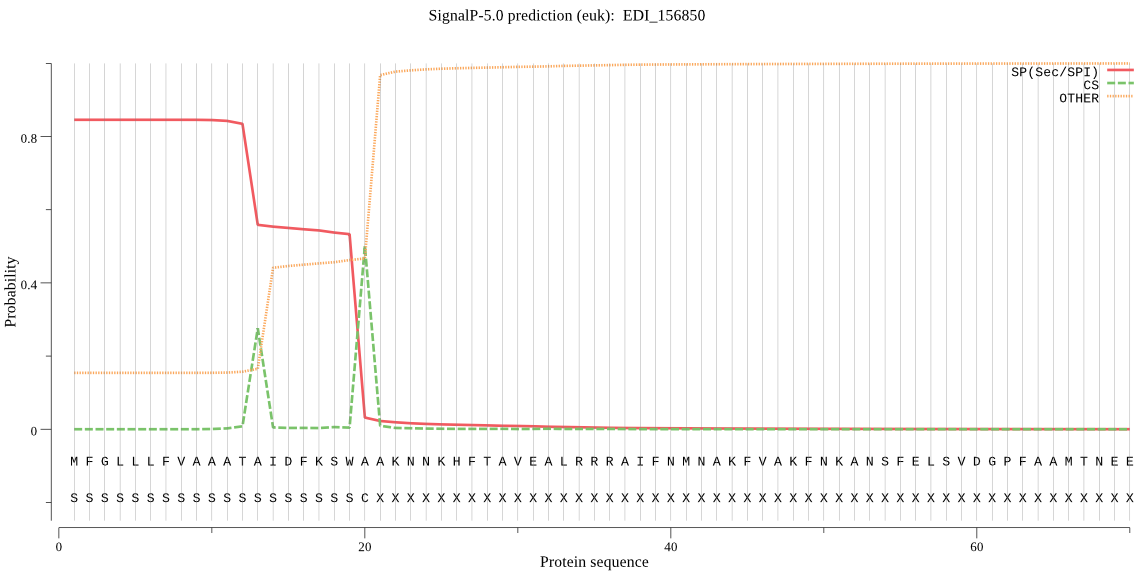
MFGLLLFVAAATAIDFKSWAAKNNKHFTAVEALRRRAIFNMNAKFVAKFNKANSFELSVD GPFAAMTNEEYNHLLRVHETEAAADSVYDNTIITASSKDWRAEGKVTPVRDQGNCGSCYS FSSLAVLESRLLIAGSKYNQNNLDLSEQQIVDCSTANNGCNGGSLSATYLYVKNNGVTDE ASYPYTATKGTCKAFTPKVKTTGLTHVTPNEDALTSALEQGPVAVCIDAGKASFQLYKSG VYDEPKCSKTVNHGVAAVGYGTQDGKDYYIVKNSWGTSWGDKGYILMSRNKNNQCAIASV AYFPNGAHDAN
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDI_156850 | T | 201 | 0.513 | 0.079 | EDI_156850 | T | 208 | 0.506 | 0.339 | EDI_156850 | T | 196 | 0.504 | 0.078 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDI_156850 | T | 201 | 0.513 | 0.079 | EDI_156850 | T | 208 | 0.506 | 0.339 | EDI_156850 | T | 196 | 0.504 | 0.078 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| EDI_156850 | 86 S | AAADSVYDN | 0.995 | unsp | EDI_156850 | 96 S | IITASSKDW | 0.997 | unsp |