

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_201400 | SP | 0.046229 | 0.888839 | 0.064932 | CS pos: 18-19. ACS-SR. Pr: 0.4189 |
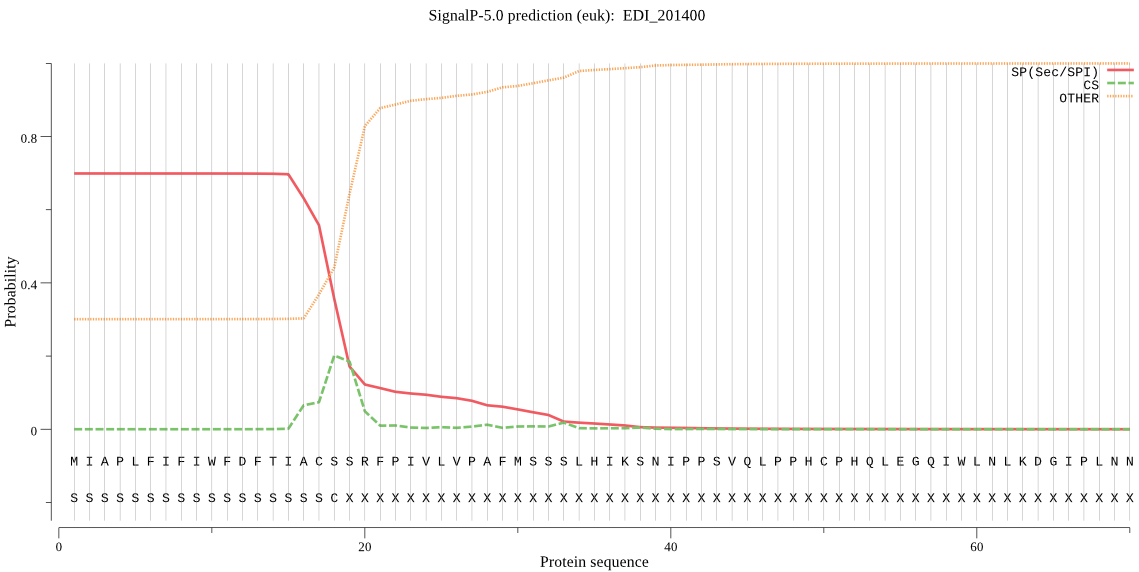
MIAPLFIFIWFDFTIACSSRFPIVLVPAFMSSSLHIKSNIPPSVQLPPHCPHQLEGQIWL NLKDGIPLNNTQCFFKYFTPFWNNSNKRFESLDGVQIYYKDFPSIEGISSLGPKEEPIVQ RVLRVWYKMIQQLKRIGYKDKKSLFGLGYDWRYADVNYNNWSKKVKEVIESAYILNNKKV MIVTHSLGGPMTLQLLFQLGNSFCEKYIEKIITISAPFIGTIKALRSFLSGETEGVPVNP LLFRDFERNIDSVYQLMPNYQWWNDTILIFNGTSYPASQMNQILNLINETKNYASFVYTN AMNRYPINWIPKVKLHCLYSSGIETEILLNYSTSFDNQPIQTFGDGDGTVSLNSLSFCKT MNLGESINIGKYDHFGIIKAQKTIDYIIEQSCKTT