

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_201690 | OTHER | 0.999661 | 0.000310 | 0.000029 |
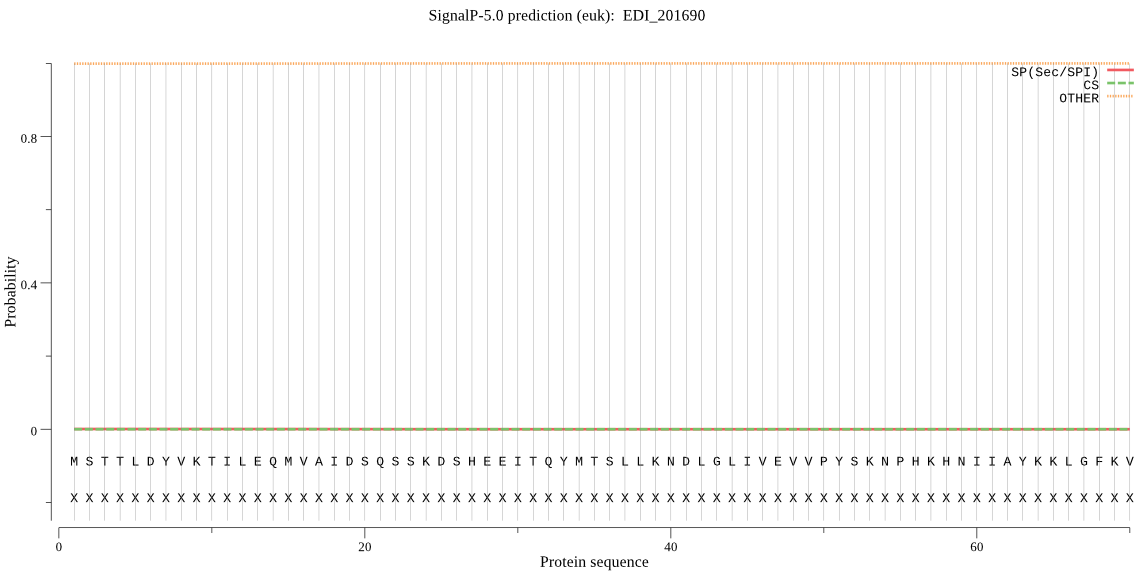
MSTTLDYVKTILEQMVAIDSQSSKDSHEEITQYMTSLLKNDLGLIVEVVPYSKNPHKHNI IAYKKLGFKVVLCGHLDTVDIGSGWTKEPLKCTTEVVDDKTYIYGRGTSDMKGGNAVIIA TLKRLIEDSNNIDDIAIFFSTEEEIGVRGCQDFMVSHKYFFESVETFVVLEPTNLYIGSG QNGHYWVKYTCHGKSANIIEAHTGVNAIEGMTDLNCVLKESITAPDLNGYVTLNIGTIEG GEKVNIVPDLCVETVYYQFGPNVFGKDLEEIIKQTIDQMNEISYAQYEYEFVRNYLQLNV DQQTSY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDI_201690 | 26 S | SSKDSHEEI | 0.997 | unsp | EDI_201690 | 26 S | SSKDSHEEI | 0.997 | unsp | EDI_201690 | 26 S | SSKDSHEEI | 0.997 | unsp | EDI_201690 | 22 S | IDSQSSKDS | 0.997 | unsp | EDI_201690 | 23 S | DSQSSKDSH | 0.992 | unsp |