

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_216570 | OTHER | 0.999896 | 0.000058 | 0.000046 |
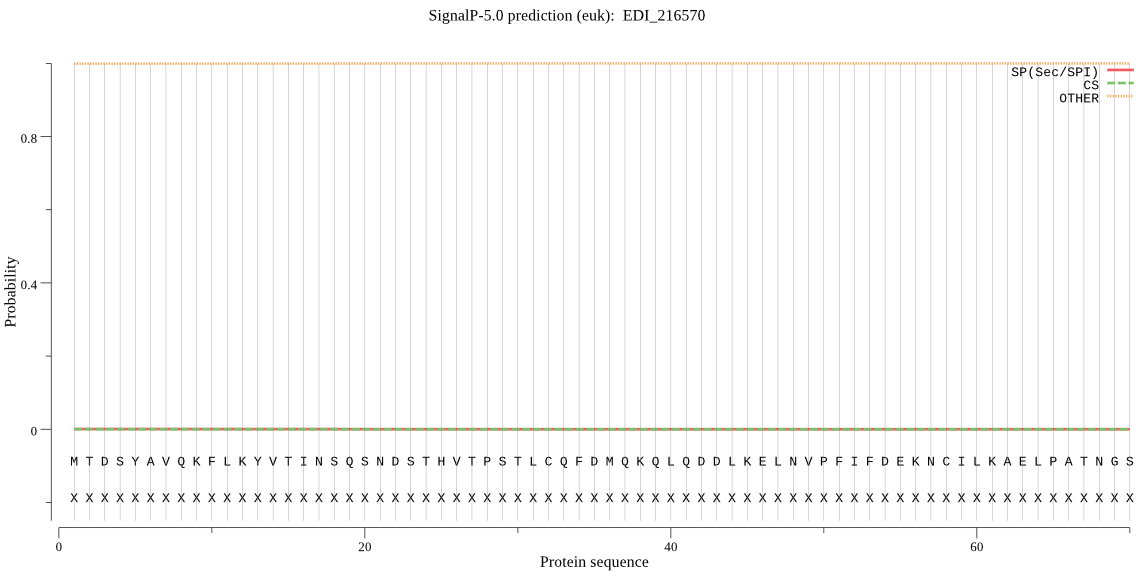
MTDSYAVQKFLKYVTINSQSNDSTHVTPSTLCQFDMQKQLQDDLKELNVPFIFDEKNCIL KAELPATNGSKQSIGFFAHIDTSPEASGEGVKPLIHQLPTEVTSDLVLPSGTVISKEDIK KYGGDEIITSSGDTLLGADDKSGVAILMGTLHKIVKDNIPHPRIMIIFTPDEEIGESCDH VSIEDLHLDYAYSIDGDELGTYNDEGFNAFGAELTINGYEVHPGEAYNIMEDAGFILSQF YTSLPISKRPETTKEDEGYILCTEMSGSVIKAHARFIVRSFKVNEMEFFIQLMKDQISFL KRKYTKSSFDLKFTEQYRNMKRYLPDNLLVDKLIQAMKLSGVEAKKQYMRGGADCSHLSE KGLPCINMFAGGMNFHSRREFIPVKAINKGVEVIGHLVQLF
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDI_216570 | 182 S | CDHVSIEDL | 0.992 | unsp | EDI_216570 | 182 S | CDHVSIEDL | 0.992 | unsp | EDI_216570 | 182 S | CDHVSIEDL | 0.992 | unsp | EDI_216570 | 252 T | KRPETTKED | 0.993 | unsp | EDI_216570 | 83 S | HIDTSPEAS | 0.993 | unsp | EDI_216570 | 115 S | GTVISKEDI | 0.995 | unsp |