

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_226130 | OTHER | 0.995789 | 0.000244 | 0.003967 |
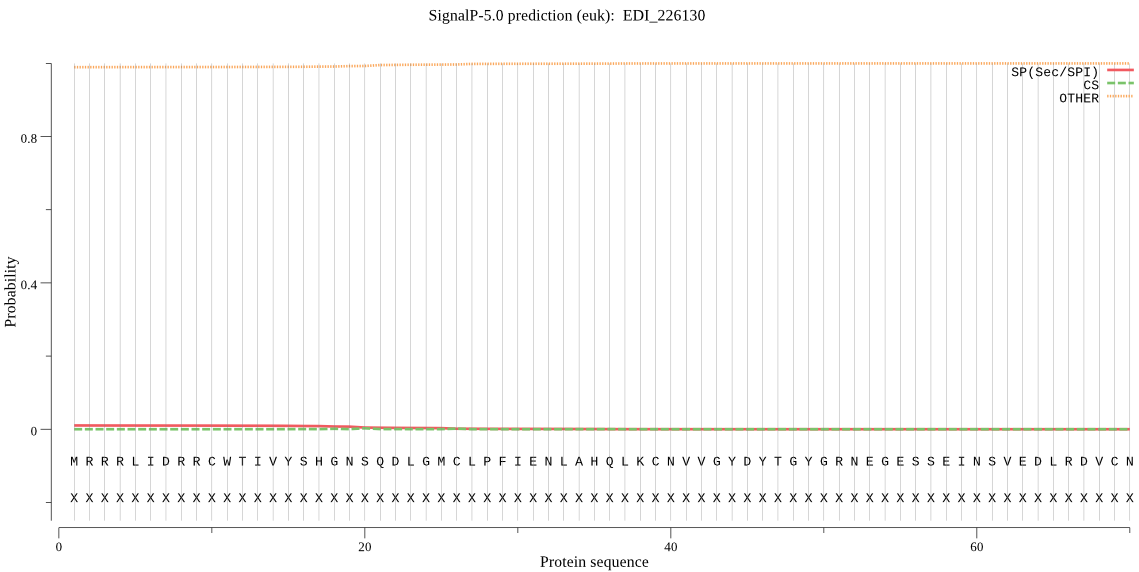
MRRRLIDRRCWTIVYSHGNSQDLGMCLPFIENLAHQLKCNVVGYDYTGYGRNEGESSEIN SVEDLRDVCNYLHNNGVSWERIVLMGHSLGGGVSISFASQECGKWEEVQEIEMNESFESK EEKKEEEIKEQKIGGMIIISTFTSICGVVSKYAGMVINDMFENITKLKHINIPVEVIHGR EDELIGVDESVEIYNSIPEEMRYGYDIINGCRHNDILENEELIKVIKRFLEKL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| EDI_226130 | 61 S | SEINSVEDL | 0.997 | unsp | EDI_226130 | 119 S | ESFESKEEK | 0.993 | unsp |