

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
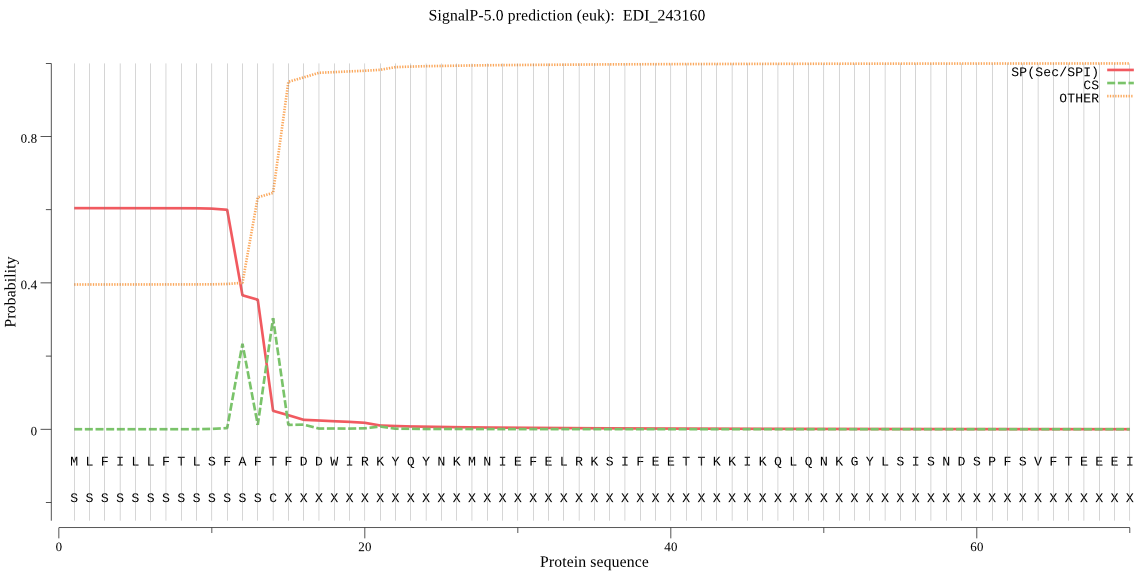
| EDI_243160 | SP | 0.067102 | 0.932859 | 0.000039 | CS pos: 14-15. AFT-FD. Pr: 0.6796 |
MLFILLFTLSFAFTFDDWIRKYQYNKMNIEFELRKSIFEETTKKIKQLQNKGYLSISNDS PFSVFTEEEIDKMINSETFSLPQLRQKRLKTNKKTIQLINEFDWRNSGIITPIKSSGTRY FEATIDGIESAILLEKKGMYTNETLKLSVQQLVDCQDEFFGKKLGRTNPCFIYLQRFGFM KEEDYPETSEKGVCQYNSSRVFGKVNKRRYLSVFNDEELIEMIKQTPIIVNIDMPSTMPY YDGEGIFENVEECSQSLPRIGLLLIGYGKTINGVPYWILKNCWGPSWGSNGYLYLKRNKN VCGIYSYGTYIESITLY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| EDI_243160 | 36 S | ELRKSIFEE | 0.995 | unsp | EDI_243160 | 212 S | RRYLSVFND | 0.993 | unsp |