

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_251670 | SP | 0.001918 | 0.998028 | 0.000054 | CS pos: 17-18. LTA-ND. Pr: 0.4737 |
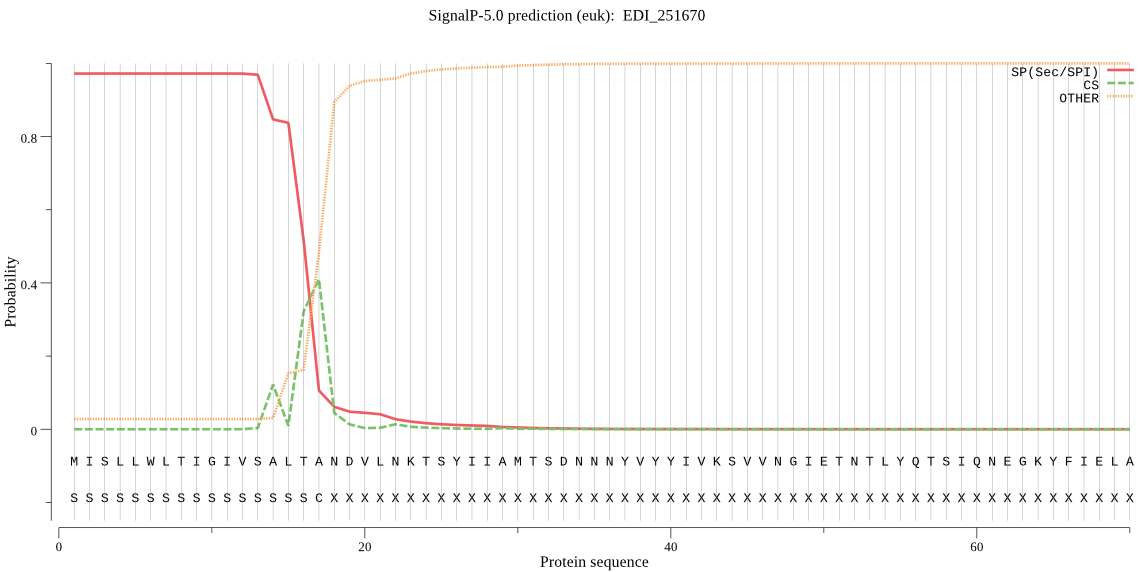
MISLLWLTIGIVSALTANDVLNKTSYIIAMTSDNNNYVYYIVKSVVNGIETNTLYQTSIQ NEGKYFIELAQGVGNELYYSVAHHSIFYTKCDSICSLYSFDSNMKEYQVTKDIDISLPVV IKEHLYFISPIFFGMTVQDSLDLQNKLVNRKQTSHHVFTQTPFGEGSSYITTNDDGTPQY KHLISSTITVDENGKINIGKYTDISNEVGNVISYDISDDETIIVFEGKSTAPASDKKNYL KSSIYTISVGSDQATCISCNYPGNQIEPKATKTGKYIIFMRTTNIIEKNPAKYFVLYDLE DGSYKADVTLNEFTTEKYLISKHNNEETISIFVINNRREALAIRKAIYSFELKTWNIEEV IVEGAVEMMAEVPCHDESSCLFITQSFQYQPSEIYYCDESMKLTKLTKLNDYTSYNLQKP ILINPMGSLNDRIQSLLYLPSNNEKAPILLVVHDEEEGTVIDGFNKIMNPYVYTNEGYAV IMINSHVSKGFGERMRIKGSNDINIIIEDIKAVINELEKYPVDTSKIAVVGVFGASKIVT ALLDAKLPIVLAAVHAPILSIRSQYYTDAKYTLEYTFEGADYDNSYWYNHNDAMKNVASY SVPTLISYGDLDFVVDKSQPISLFQALQRRGIKSRLLRFPYSTNVLTKNEDIKVFYEQYL RWFNENMKN