

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_252380 | SP | 0.057143 | 0.942788 | 0.000069 | CS pos: 17-18. CFG-DE. Pr: 0.9580 |
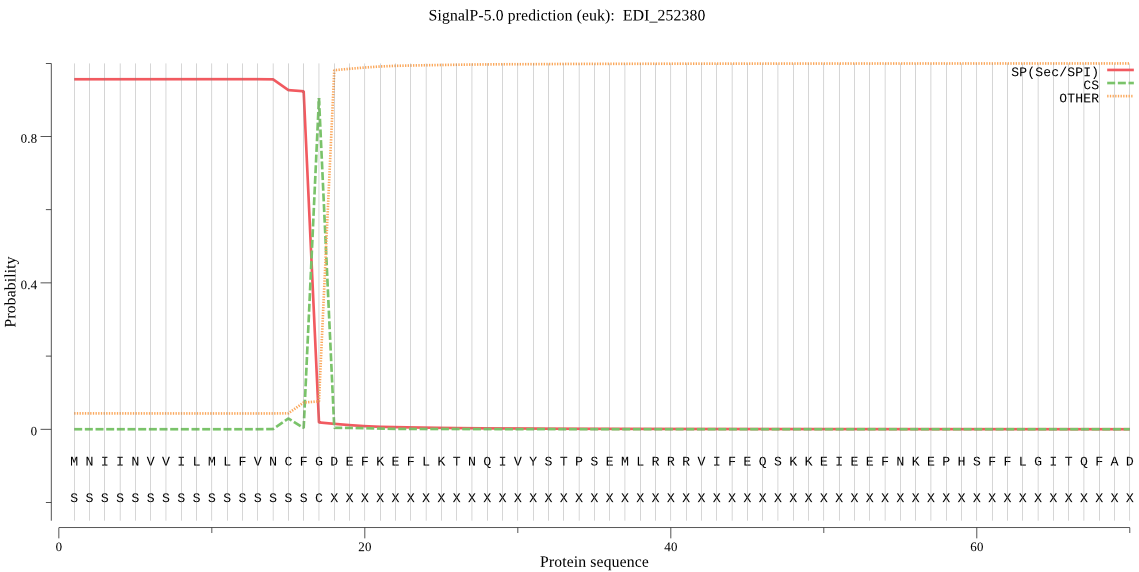
MNIINVVILMLFVNCFGDEFKEFLKTNQIVYSTPSEMLRRRVIFEQSKKEIEEFNKEPHS FFLGITQFADKTDEEFNQMFSMKMDRQDEFSMMSTDEEENKNESDDIYMEYNLNQDEETY GREMRRKVRGKKYRGVRFMRKVHVPKKYRIGRKWQFNKKKDSVKELPEGIDFRKFGKLTY IREQTGCGGCWSFASVCALESRYLIDYNLTVDDVGRTWALSEQQLLDCCIENNGCEGGSM ERSFRCMNRTKGVMQRVRYPYEAETQDCKEFNNEYKEVTLGGYALVPRGNERALMSAIHK FGVLGIGLDTRSKLFKHYRGGIYYNEECTRRGLSHAMNLVGYGTTKEGQKYYIIRNSWGD WKWGEDGYMRLYRGGNHCGVATNAFFPLFVRRVNDEPKPMSHTETMIKDLINRHRLGWI
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDI_252380 | 401 S | PKPMSHTET | 0.995 | unsp | EDI_252380 | 401 S | PKPMSHTET | 0.995 | unsp | EDI_252380 | 401 S | PKPMSHTET | 0.995 | unsp | EDI_252380 | 162 S | KKKDSVKEL | 0.998 | unsp | EDI_252380 | 357 S | IIRNSWGDW | 0.997 | unsp |