

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_276390 | OTHER | 0.981761 | 0.017973 | 0.000267 |
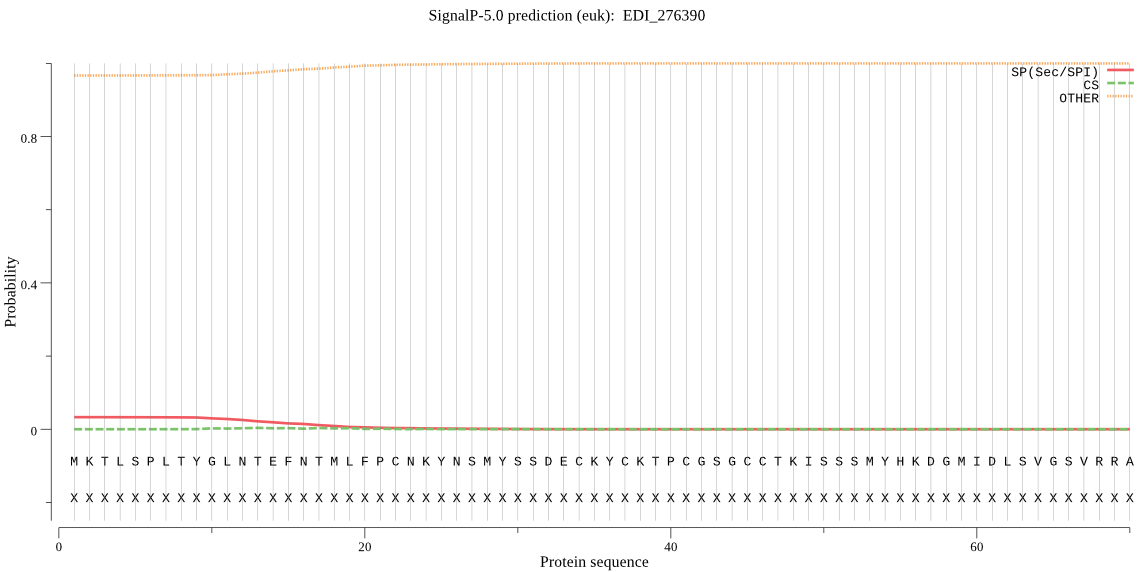
MKTLSPLTYGLNTEFNTMLFPCNKYNSMYSSDECKYCKTPCGSGCCTKISSSMYHKDGMI DLSVGSVRRAGHSMLVVGWNDEFPVERNRYSSPPIYTAAWNAIDNKTQTKLTKNNVEIPL DRWTKGGLILKNSYGEHGHTLGYFLQNHSLLNEDTICPLSIHPSRYYPLDVECIKSGNSF MNCSNDKLRKVSDGKIFYGATILKCKSELGNDYAKNLGFEECGTNKDYRYALSMEWFNVT FKRGVCKTREMGDGLIVYLARWVEGKMKETITEVHTQITSWDFIAQLFEMDTTVKNSEHC GYYFQPYDTIEYLISMYQHDGFESYGFSYYGIKWEQESYYKSSKRKGQFTEINKSTKKFV TPQFDGPFGGRIN
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EDI_276390 | 355 S | EINKSTKKF | 0.992 | unsp |