

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_280180 | SP | 0.008772 | 0.991183 | 0.000045 | CS pos: 14-15. CFG-IE. Pr: 0.4181 |
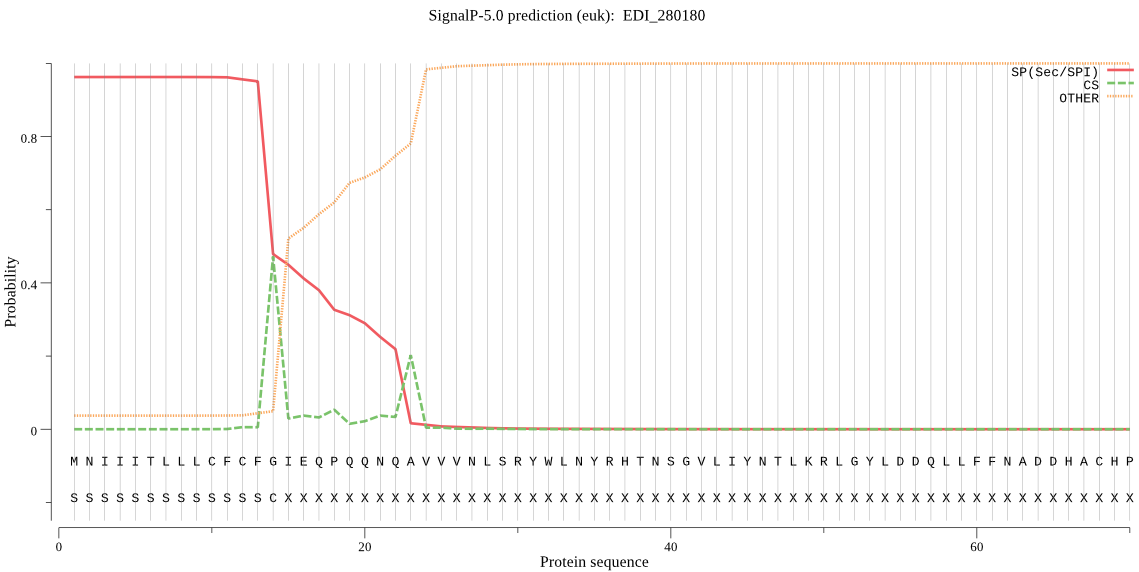
MNIIITLLLCFCFGIEQPQQNQAVVVNLSRYWLNYRHTNSGVLIYNTLKRLGYLDDQLLF FNADDHACHPRNIFPGEMRLNTNMPNIYKDIIIDYKGRDVSIEKYMRAMLGRDVKGTPDS LRLVRGERTFIYLIGHGGEGFMKFQNRDEITSYDIEYMFKEMEIMKRYKEVMFIVDTCQA TSLSDRIKAKNIITVGSSVTGQSSYSGYISNEIGAITSDLWDQHQDVLFQRSLNKESNMT VQDYLNYFNKNMLKSNHGWRSDLFNRPLDQVKMTDFLAYIPQSVDVNLNEIPWSDNIFN