

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_284230 | SP | 0.082394 | 0.917233 | 0.000373 | CS pos: 13-14. AYA-SL. Pr: 0.3619 |
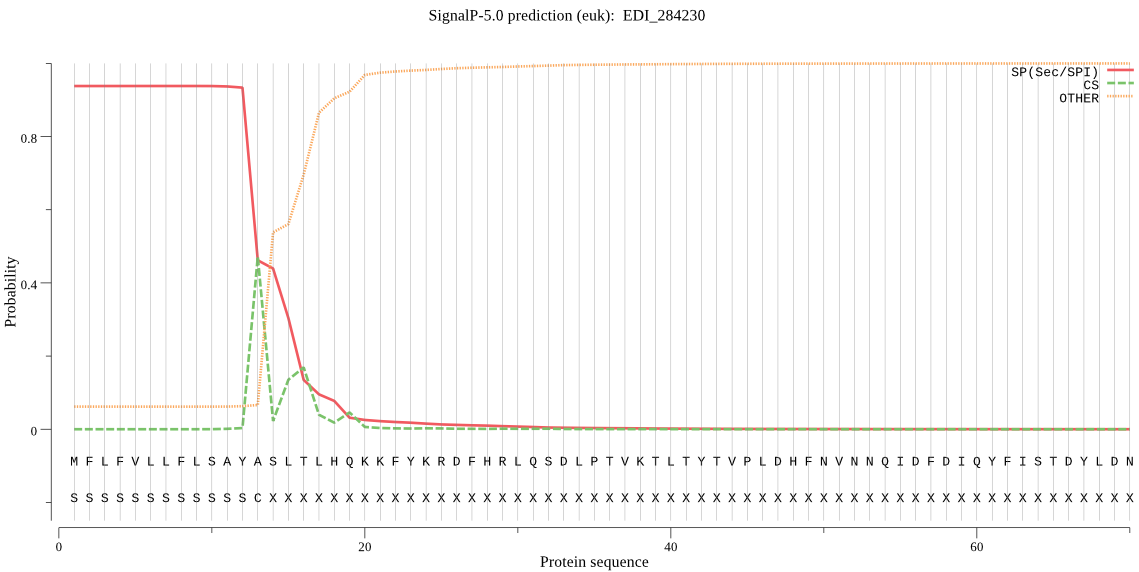
MFLFVLLFLSAYASLTLHQKKFYKRDFHRLQSDLPTVKTLTYTVPLDHFNVNNQIDFDIQ YFISTDYLDNNSPNAPLFVLLGGEGPEDATGLQNYFVVTDLAKKHKGLMLSVEHRFYGAS TPSLEMDKLIYCTAEQALMDYVEVISHVQEENNLVGHPVIVLGGSYSGNLAAWMRQKYPN VVEGAWASSAPVEAVVDFYQYLEVVQNALPKNTADLLSFAFEKWDEMTTTEEGRKELGKI FNTCTEFGEKDIQTFAESIGTALSGYVQYNSSNWKSSYESTDSICTEINEDVVNKYPLFI KEKYNPEWGDKECTSSSQEESYKTLQSTSTYAEGNEGAAGRSWFFQTCIAYGYYQAVSEQ SSVKWGKLNQLQGSIDMCKDIYGIDKDTLYNAVDHINVRYGGKKPCVTNVAFTNGNTDPW HALGVTESDHQEGNLVQLIDRTSHCSDLYSEKETDVPELKKARHNELKFIAQVLANVPQN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDI_284230 | 362 S | SEQSSVKWG | 0.995 | unsp | EDI_284230 | 362 S | SEQSSVKWG | 0.995 | unsp | EDI_284230 | 362 S | SEQSSVKWG | 0.995 | unsp | EDI_284230 | 450 S | SDLYSEKET | 0.996 | unsp | EDI_284230 | 277 S | NWKSSYEST | 0.996 | unsp | EDI_284230 | 317 S | CTSSSQEES | 0.996 | unsp |