

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
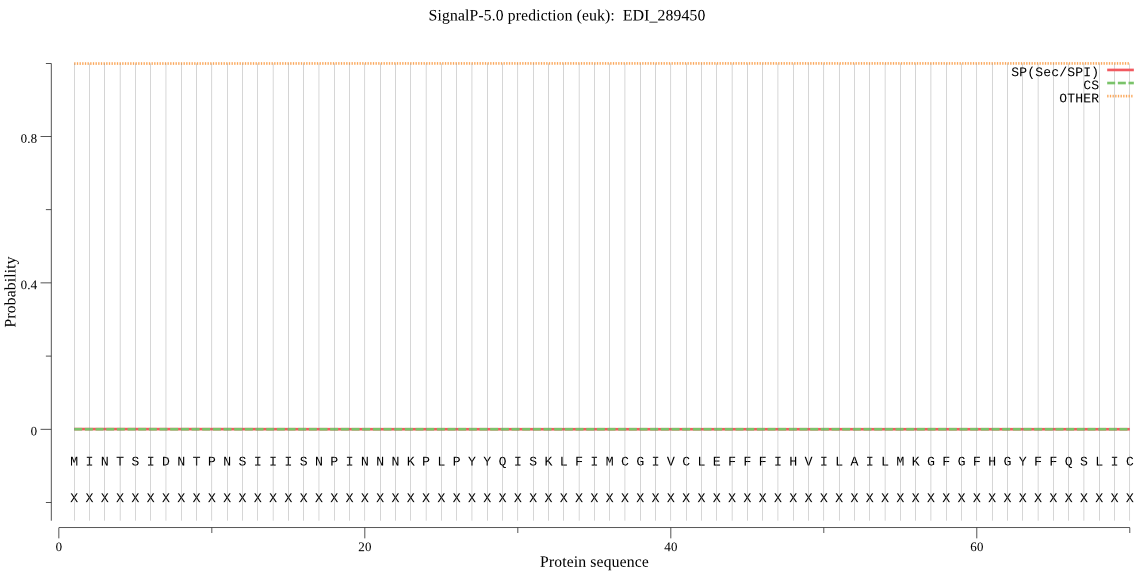
| EDI_289450 | OTHER | 0.999997 | 0.000000 | 0.000002 |
MINTSIDNTPNSIIISNPINNNKPLPYYQISKLFIMCGIVCLEFFFIHVILAILMKGFGF HGYFFQSLICSLIVINLMYYRYSMNKNIITLNTFIQEILKFNKNSFPTLIIALLGLICIL FLRIFLYYCHIDTSDLFTNFYTDGQLDIPIIIDTLFFSVITEEIALRRSMLNTIKIYTQN PKNYLFIISGVWFGLIHFINIGTSSFTVPYVFFQMSYGMIIGYTFSIFTESLGIFPTILL HSMNNALAIINPFVFSPFLLLYATLSALLFIYLSHKLLITHSL
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| EDI_289450 | T | 4 | 0.521 | 0.031 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| EDI_289450 | T | 4 | 0.521 | 0.031 |