

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_289460 | OTHER | 0.999907 | 0.000051 | 0.000042 |
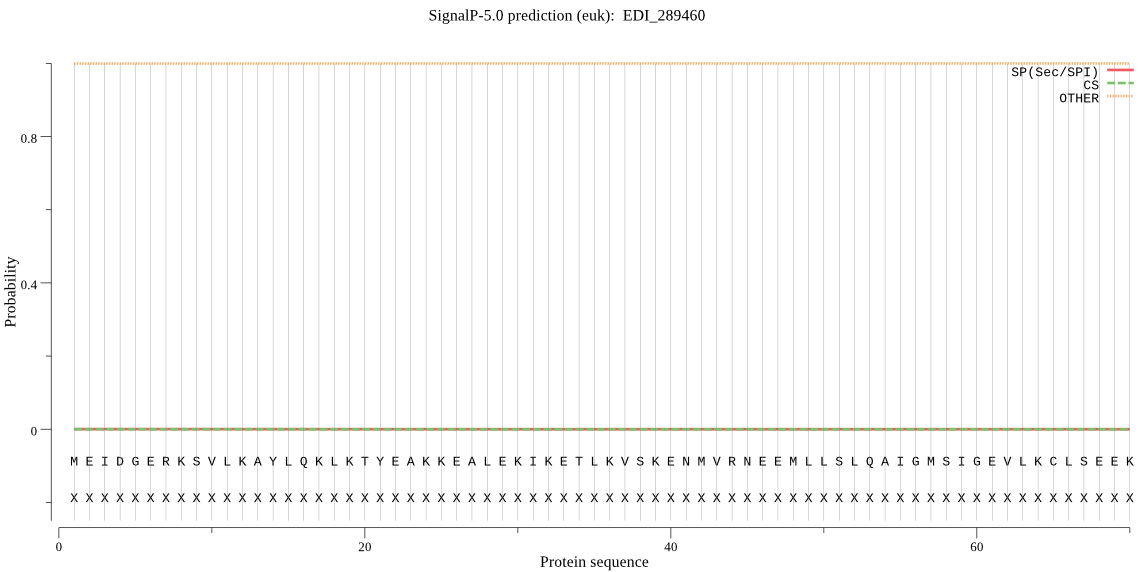
MEIDGERKSVLKAYLQKLKTYEAKKEALEKIKETLKVSKENMVRNEEMLLSLQAIGMSIG EVLKCLSEEKYIVKVSLGPHYVVGCRNSIDKKKITQGTRVALDVSTNTIMKILPREVHPG VYSMTVESPGKVKYEDIGGLRNQMREIREVIELPMTNPELFERVGVKAPKGVLLYGPPGT GKTLLARALASNLECQFLKVVASGIVDKYLGESARLIREMFAYARDHQPCVIFMDEIDAI AGKRIAEGIHSDREIQRTLMELLAQMDGFNEISKVKIIMATNRPDVLDPALMRPGRLDRK IEISLPNDQGRIEILKIHSKKMNIKGEIDYEAMGKLTEGFNGADLRNVCTEAGMFAIRDD RDYCLEEDFFKAIRKQSDAKKLENQFGDIKI