

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_299030 | OTHER | 0.999999 | 0.000001 | 0.000000 |
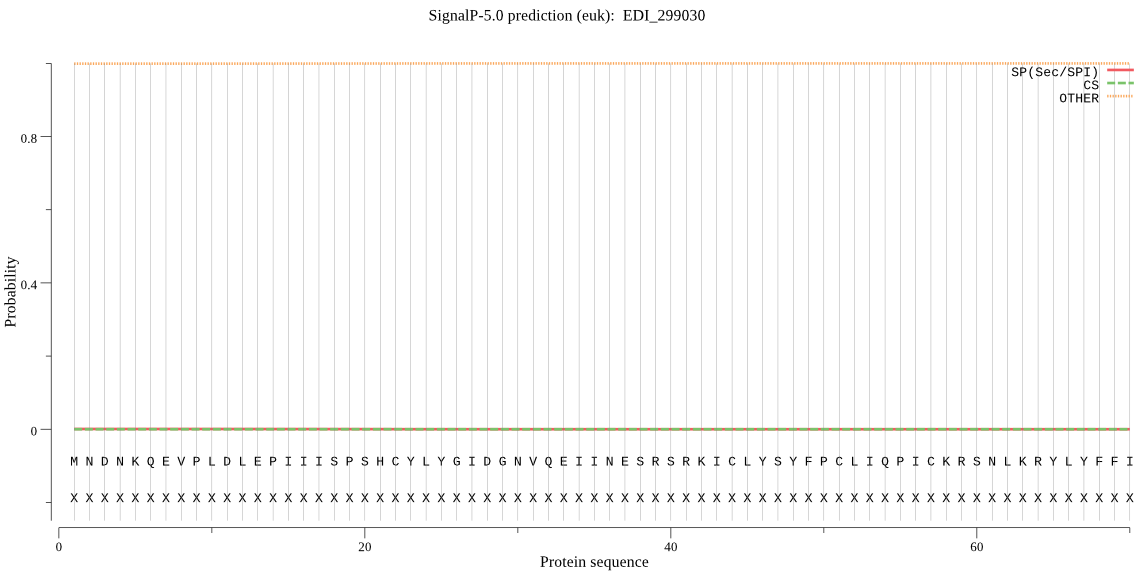
MNDNKQEVPLDLEPIIISPSHCYLYGIDGNVQEIINESRSRKICLYSYFPCLIQPICKRS NLKRYLYFFISVTFIITTIELILFFISALFEHEKDSYILILTTNSIKIVGGKNLSLIKCE YQYFRLVLPIFLQNNLLQLIVTTFIQLKFGIIVERILGSSWYFIMYLFCGTMPLFGSYLF AYKLPFIGCSSSLIGIMIIWILQMIIQKEKYHPIVWVQLFISAIFSVLLIIGSCLFTQID GVSIIFSIGTGMLCGLTYFGDNFKSCLTNIFFKWANRLLPIILYLSILTLTIFVIHFVLP VSCHSTQTAS
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDI_299030 | T | 308 | 0.543 | 0.033 | EDI_299030 | T | 306 | 0.506 | 0.038 | EDI_299030 | S | 310 | 0.506 | 0.014 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDI_299030 | T | 308 | 0.543 | 0.033 | EDI_299030 | T | 306 | 0.506 | 0.038 | EDI_299030 | S | 310 | 0.506 | 0.014 |