

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
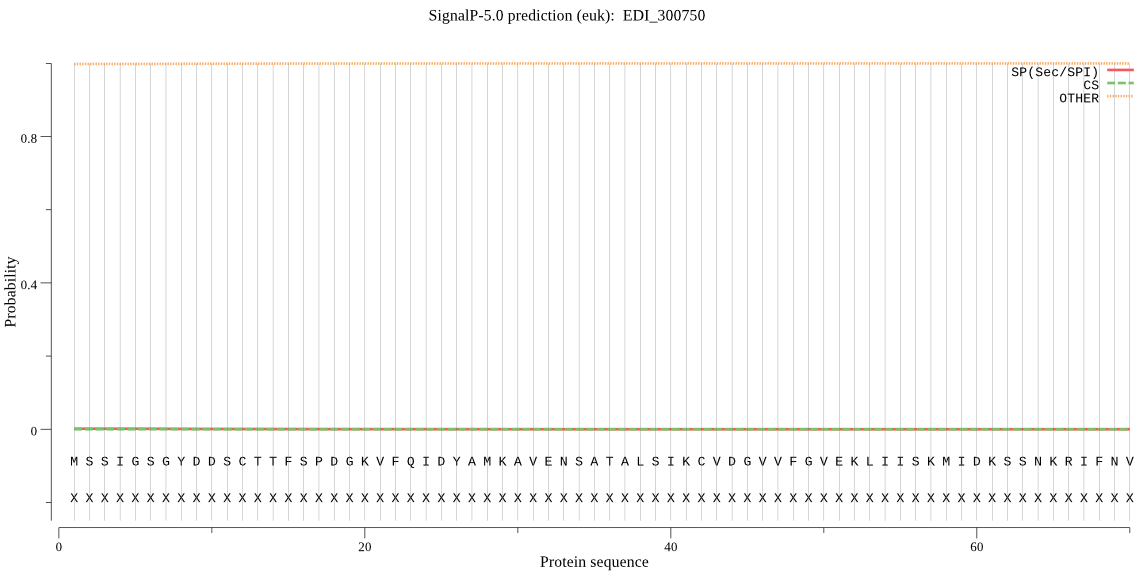
| EDI_300750 | OTHER | 0.999945 | 0.000053 | 0.000002 |
MSSIGSGYDDSCTTFSPDGKVFQIDYAMKAVENSATALSIKCVDGVVFGVEKLIISKMID KSSNKRIFNVGTHIGITCAGFLPDARKLAKKAQEEAESFKENYGMNIPVKVLAEKIAMYM QNYTLYGSLRPFGCSLLIGGVDQHGYSLHYIEPSGTTYGYFANGIGKGKQICKNELEKIN FSQITCEEALIKLVEIIDLCHSEWKDKPYEIELSWISEKTNNIVKKVDKEMKQKIIDQAR NKQMHQN
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| EDI_300750 | S | 2 | 0.522 | 0.053 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| EDI_300750 | S | 2 | 0.522 | 0.053 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| EDI_300750 | 6 S | SSIGSGYDD | 0.993 | unsp | EDI_300750 | 98 S | EEAESFKEN | 0.996 | unsp |