

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EDI_315330 | SP | 0.015449 | 0.984209 | 0.000342 | CS pos: 15-16. VDT-SS. Pr: 0.3949 |
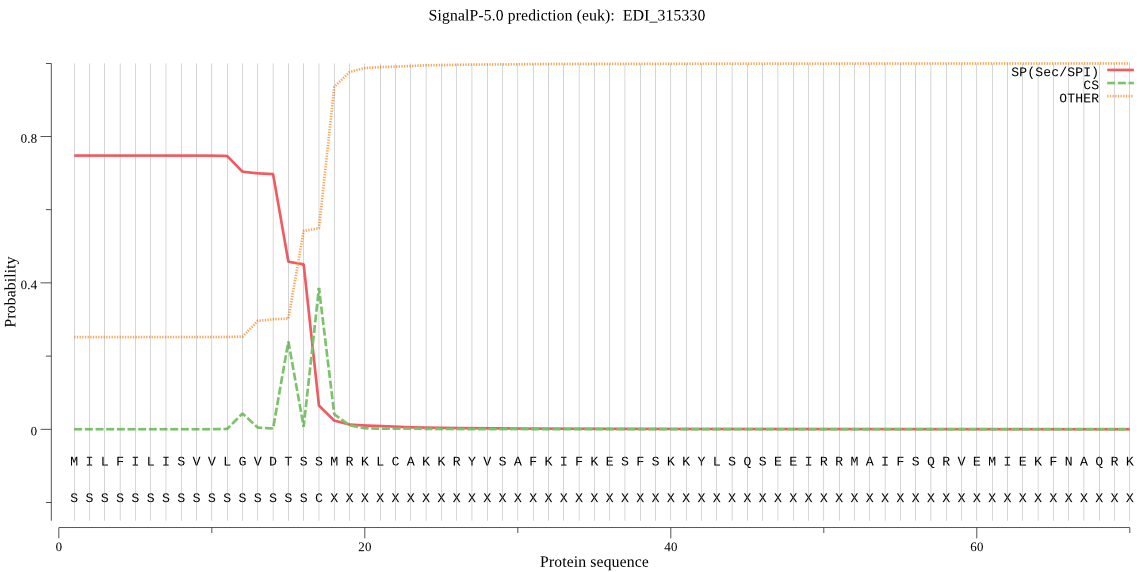
MILFILISVVLGVDTSSMRKLCAKKRYVSAFKIFKESFSKKYLSQSEEIRRMAIFSQRVE MIEKFNAQRKIESDVKLDINNFTDLTNDEFIQMYIGGKKKSENSKDQNIVNNNRYIPQKV NYPTMYSLCGKNTNYNSETDGKIDHCSLGVDQKLCRCCYAASIANFLQIKHHIANGENVQ YSIQQIVDCTGGKTKGCCSGMSYDALNYNRVFATNDVYPFRDAETSPDCQSYTRPSCYTV KEFAQTLTSYKEWYAPTYNEIKQIIYDNKGAISGIYVPLSGVVSEVWQSYSSGIINVSSY CSSNNGNIFTNHMVVLVGFGFEAQGEGVNGNFIIIRNSWGTDWGDKGTAKLSTDSLCGIG NCDGETVPCSHPTIITFETKVGPSTEQDYGCLQDKCSPTNDCDGVNCLTKSLDPPPNHSV GIVIIMLVICLMGLF
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDI_315330 | 384 S | KVGPSTEQD | 0.994 | unsp | EDI_315330 | 384 S | KVGPSTEQD | 0.994 | unsp | EDI_315330 | 384 S | KVGPSTEQD | 0.994 | unsp | EDI_315330 | 44 S | KKYLSQSEE | 0.996 | unsp | EDI_315330 | 249 S | QTLTSYKEW | 0.996 | unsp |