

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_011810 | SP | 0.001205 | 0.998787 | 0.000008 | CS pos: 18-19. SYA-LY. Pr: 0.9332 |
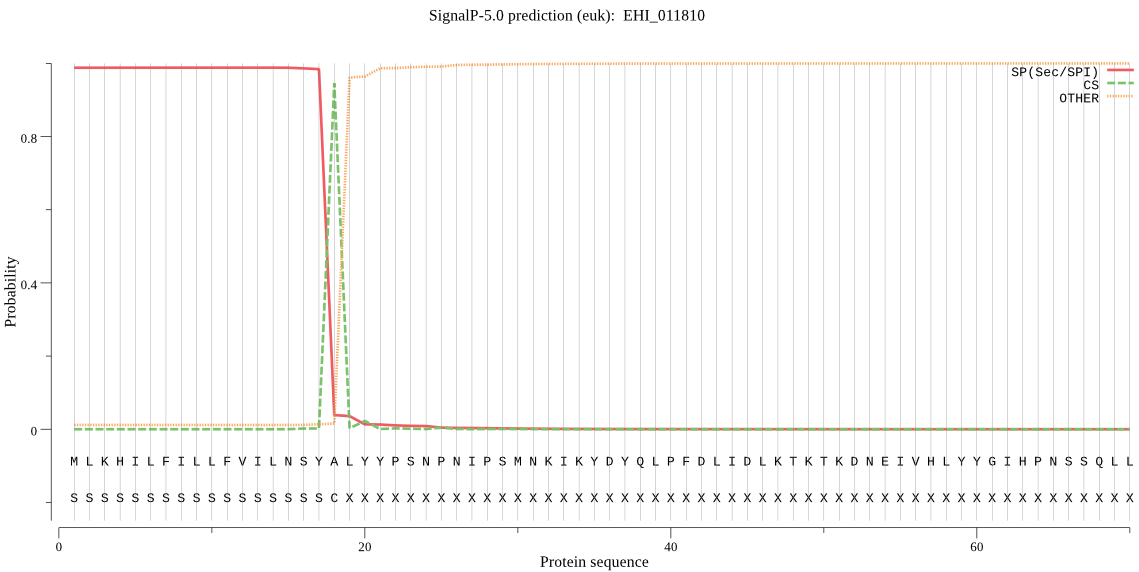
MLKHILFILLFVILNSYALYYPSNPNIPSMNKIKYDYQLPFDLIDLKTKTKDNEIVHLYY GIHPNSSQLLVLFQSNAGCFLDRLFLLKSFYSKFNISVGILSYRGYGNSTGKPSEQGFIE DALASLSHLSKDGIPIQNITIIGRSIGVGVALSVAQILPIKKLILENGFTNLVDFLPNLQ NNEVMIRDPWLNEQKIETINKKTSILFLLSEGDEIVPPWMTRKMEMKARSMGIQTKLVSF PGARHMQLPYYDNYYRVIKEFLIEQNQGDFSK