

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_021300 | SP | 0.028223 | 0.970492 | 0.001285 | CS pos: 16-17. TIA-CS. Pr: 0.5692 |
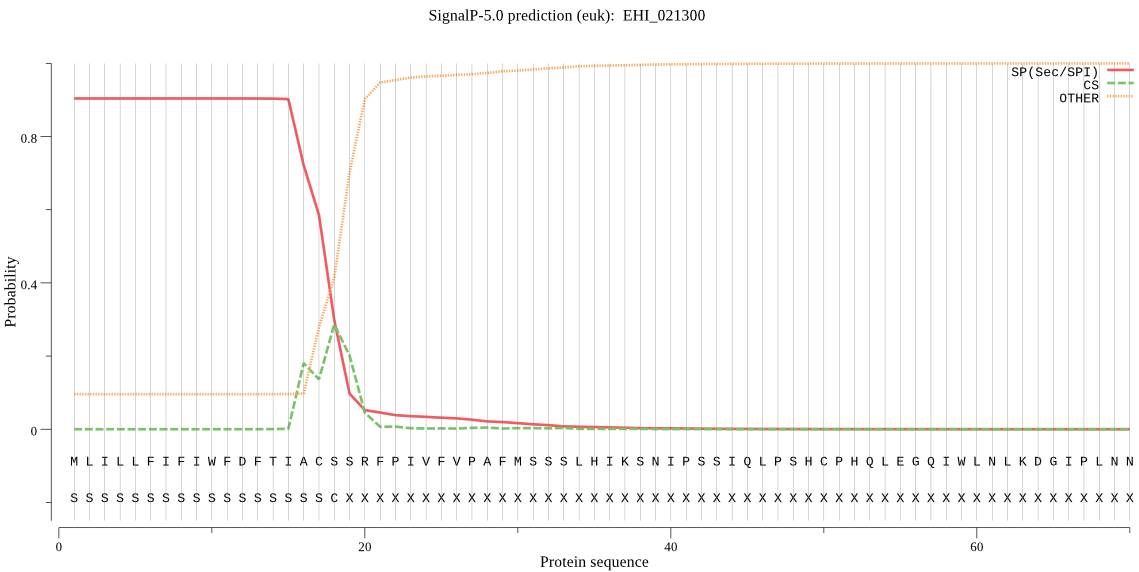
MLILLFIFIWFDFTIACSSRFPIVFVPAFMSSSLHIKSNIPSSIQLPSHCPHQLEGQIWL NLKDGIPLNNTQCFFKYFTPFWNNSNKHFESLDGVQIYYKDFPSIEGISSLGPKEEPIVQ RVLRVWYKMIQQLKRIGYKDKKSLFGLGYDWRYADVNYNNWSKKVKEVIESAYILNNKKV MIVTHSLGGPMALQLLFQLGDSFCEKYIEKIITISAPFIGTIKALRSFLSGETEGIPVNP LSFRNFERNIDSVYQLMPNYQWWNDTILIFNGTSYSASQMNQILNLINETKDYASFIYTN AMNRYPINWTPKVKLYCLYSSGIETEVLLNYSTSFDNQPIQTFGDGDGTVPLNSLSFCKT MNLEESINIGKYDHFGIIKAQKTIDYVIEQSCKTT