

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
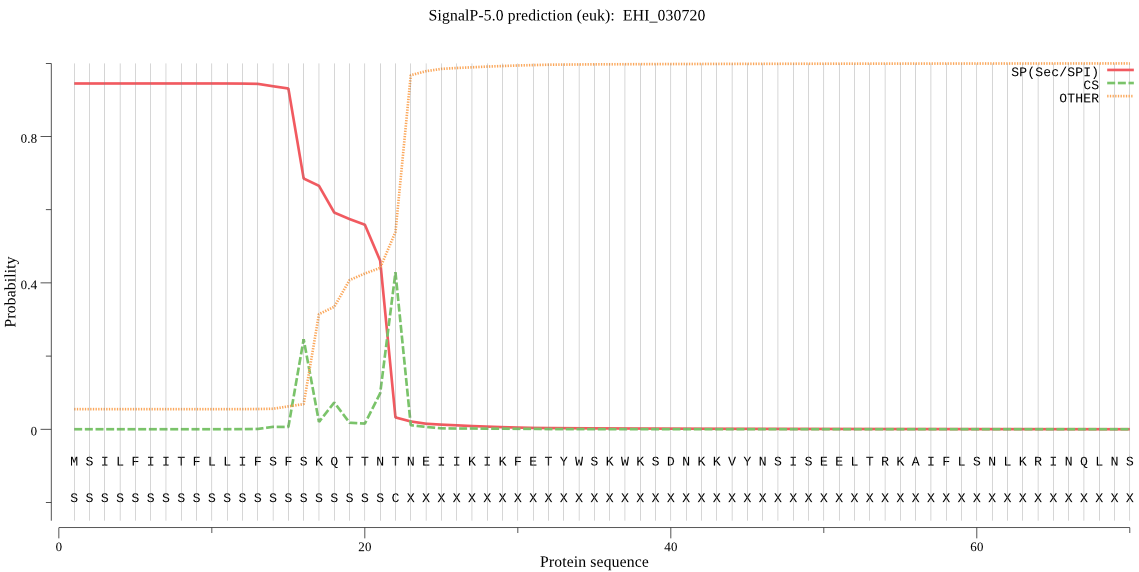
| EHI_030720 | SP | 0.009899 | 0.990017 | 0.000085 | CS pos: 16-17. SFS-KQ. Pr: 0.2630 |
MSILFIITFLLIFSFSKQTTNTNEIIKIKFETYWSKWKSDNKKVYNSISEELTRKAIFLS NLKRINQLNSQRIDTDDAVFGINAFSDLKPEEFARRFNKINLKSLKPKQTTHYKLPVPSG EVPTQYSACLQNKLLGQNSSNNIDLCGGIVMDQGDCGNCYAVSNAHELQLKYANLTLTQR GKVEYEMFSPQQLMDCTENSYYCEGGTSDEPLMSSHYVVFEKDYPYISYSNTLVNNSCIH DKPTPMMVSFSLFDSAQNFEVLKRIIYHYGSFITSVKASSDWLYYHSGIYSHSCTKNVIT NHVIEVIGYGNQNGKEYLIARNSWGKNWGIDGFIKISAKSLCGIGGDDGGMYPVSLVQHA DFSDIPKGVYGKKSYKQEVQPKLYNGTDKYDIYEQFPEVSGNSFIDIIINFIVTIALYVG LICWVICVIVILVMSCVVIYLKKN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| EHI_030720 | 47 S | KVYNSISEE | 0.991 | unsp | EHI_030720 | 374 S | YGKKSYKQE | 0.99 | unsp |