

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
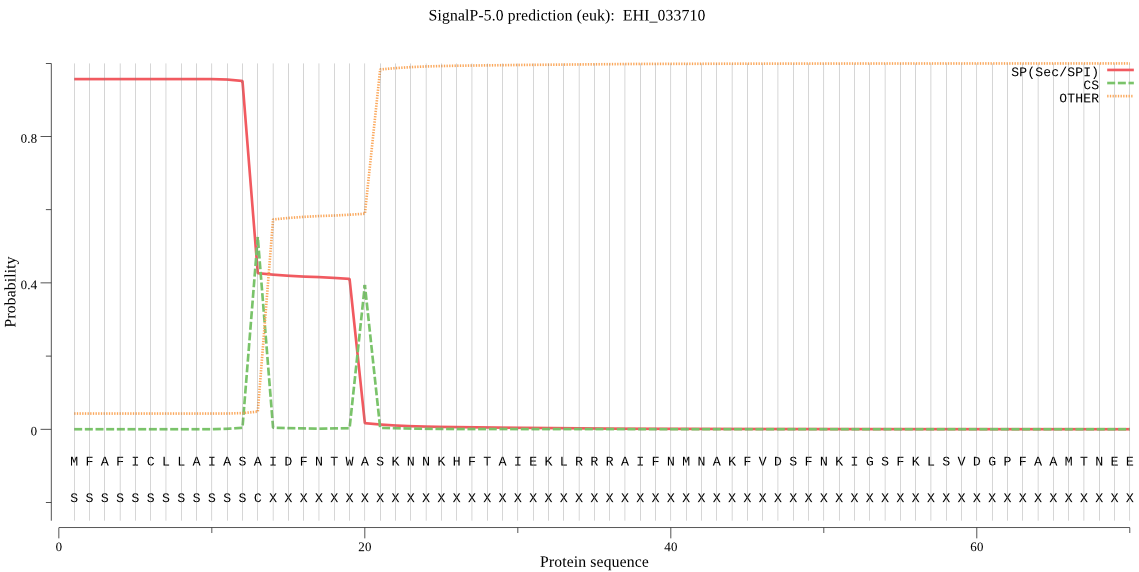
| EHI_033710 | SP | 0.011438 | 0.988312 | 0.000250 | CS pos: 13-14. ASA-ID. Pr: 0.5773 |
MFAFICLLAIASAIDFNTWASKNNKHFTAIEKLRRRAIFNMNAKFVDSFNKIGSFKLSVD GPFAAMTNEEYRTLLKSKRTTEENGQVKYLNIQAPESVDWRKEGKVTPIRDQAQCGSCYT FGSLAALEGRLLIEKGGDANTLDLSEEHMVQCTRDNGNNGCNGGLGSNVYDYIIEHGVAK ESDYPYTGSDSTCKTNVKSFAKITGYTKVPRNNEAELKAALSQGLVDVSIDASSAKFQLY KSGAYTDTKCKNNYFALNHEVCAVGYGVVDGKECWIVRNSWGTGWGDKGYINMVIEGNTC GVATDPLYPTGVQYL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EHI_033710 | 77 S | TLLKSKRTT | 0.991 | unsp |