

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_035750 | SP | 0.009316 | 0.990400 | 0.000284 | CS pos: 13-14. TWG-KV. Pr: 0.4767 |
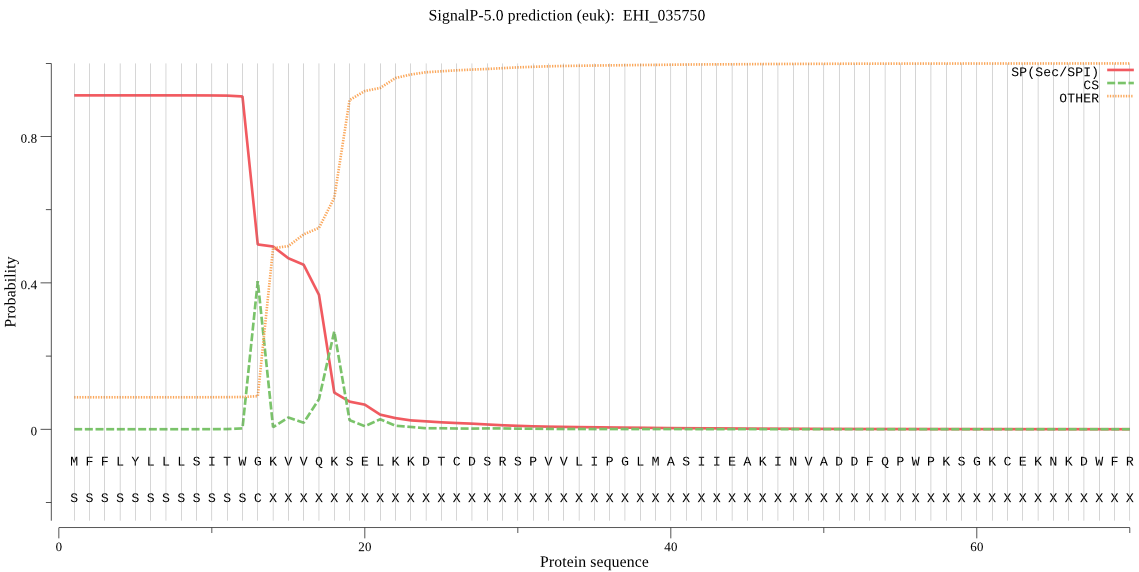
MFFLYLLLSITWGKVVQKSELKKDTCDSRSPVVLIPGLMASIIEAKINVADDFQPWPKSG KCEKNKDWFRAWVNVDIAVPWKSECYINYLSGIWNNQTNKLETIPGIDLRIPEFGSTYAC DQLDPVFLIGSFTNSFHKIIEHLKSVGYKDQIDMFGASYDWRTVDLPKTYFEGVKGLIYE GFKNSGKKVVIISHSMGGFVSYKLFDYLGKDFCDKYIQKWIAISAPFIGTGVVPKQMTVG ENLGLPIKAEYARDLSRSIESVLALSPNEEKWNDDILVRIKSNGKTYTAKQLREVYKQIP ELKDKTDYILDTEMTPLYKKWNWTIPNGVKMDCVYSHGKETPYSIEFDTEDLTKGYTVNY SDGDNLVNINSLESCKIFTDSVTNLGKHGHLLILNSNDIWNYIKPKVCGNYD
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EHI_035750 | 266 S | VLALSPNEE | 0.995 | unsp |