

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_037190 | SP | 0.085567 | 0.909552 | 0.004882 | CS pos: 13-14. AYA-SL. Pr: 0.3665 |
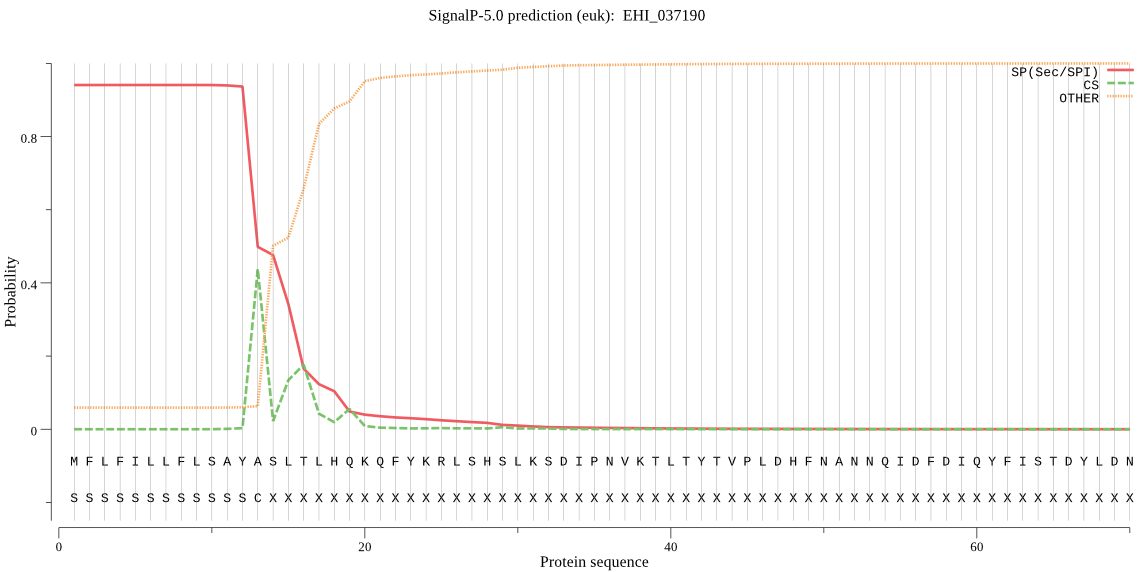
MFLFILLFLSAYASLTLHQKQFYKRLSHSLKSDIPNVKTLTYTVPLDHFNANNQIDFDIQ YFISTDYLDNNSPNAPLFVLLGGEGPEDETGLQNYFVVTDLAKKHKGLMLSVEHRFYGAS TPSLEMDKLIYCTAEQALMDYVEVISHVQEENNLVGHPVIVLGGSYSGNLAAWMRQKYPN VVEGAWASSAPVEAVVDFYQYLEVVQNALPKNTADLLSFAFEQWDKMTTTEEGRKELGKI FNTCTEFGEKDIQTFAESIGTALSGYVQYNSSNWKPSYESTDSICAEINEDIVNKYPLFI KEKYNPEWADKECTPSSQEESYKTLQNTSTYAEGNEDASGRSWFFQTCIAYGYYQAVSEQ SSVKWGKLNQLQGSIDMCKDIYGIDKDTLYNAVDHINVRYGGKKPCVTNVAFTNGNTDPW HALGVTESDHQEGNLVQLIDRTSHCSDLYSEKENDVPELKKARHNELKFFAQVLANVPQN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EHI_037190 | 317 S | CTPSSQEES | 0.996 | unsp | EHI_037190 | 317 S | CTPSSQEES | 0.996 | unsp | EHI_037190 | 317 S | CTPSSQEES | 0.996 | unsp | EHI_037190 | 362 S | SEQSSVKWG | 0.995 | unsp | EHI_037190 | 450 S | SDLYSEKEN | 0.995 | unsp | EHI_037190 | 29 S | RLSHSLKSD | 0.993 | unsp | EHI_037190 | 277 S | NWKPSYEST | 0.996 | unsp |