

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_037210 | OTHER | 0.574020 | 0.423829 | 0.002150 |
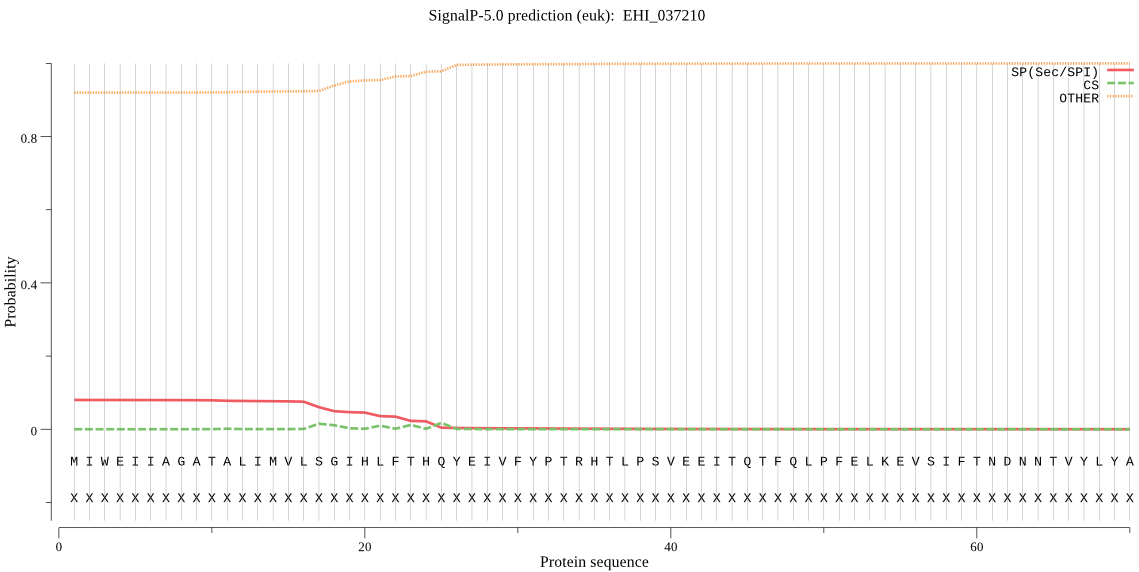
MIWEIIAGATALIMVLSGIHLFTHQYEIVFYPTRHTLPSVEEITQTFQLPFELKEVSIFT NDNNTVYLYACLKEEPSKHITLLLFQSNAGSVLDRMEMAKKYYELCNVNFVIAVYRGFDK STGIPEEVTMNNDVEKYFDSLESLGVDMNNIVVIGRSIGASMALKLYNKKNCKGLIIENG FTTLLDVGKILMPAISFFPWLIKDKWDNLNEIKHVQKGKRILFCSSGQDEMVPPSMMQHL YDVAHETGKKVRMEKFAKGFHMNLPSYPEYFKKLNKFFEELTKETMEEGIIENQEVQ
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EHI_037210 | 39 S | HTLPSVEEI | 0.995 | unsp |