

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_039610 | SP | 0.142370 | 0.857311 | 0.000319 | CS pos: 13-14. VNG-ID. Pr: 0.8846 |
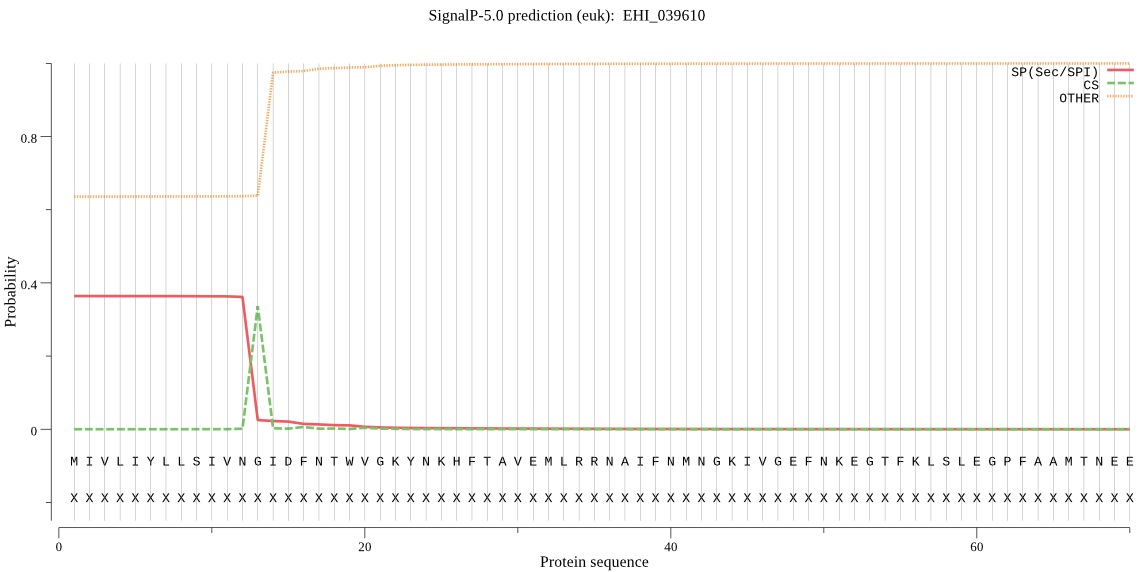
MIVLIYLLSIVNGIDFNTWVGKYNKHFTAVEMLRRNAIFNMNGKIVGEFNKEGTFKLSLE GPFAAMTNEEYKNILKSKRSEEGKGKVKYLNIEAPETVDWRKEGKVTPIRDQAECGGCYA FGSIAALEGRLLVEQGGDANTLDLSEEEMIQCTREYGNNGCGGGFGSNAYDYIIEHGVSN ETEYPFTGMDSECKTNIKPYVTMKGYNRVARNNVKELKSAISQGMVDVAMDASSVKFQLY KSGAYTDKKCKKSFFALNHEVSAVGYGVVDGIECWIIRNSWGTTWGENGYFNIAIEGNTC GIATDPLYPTGAQYL