

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
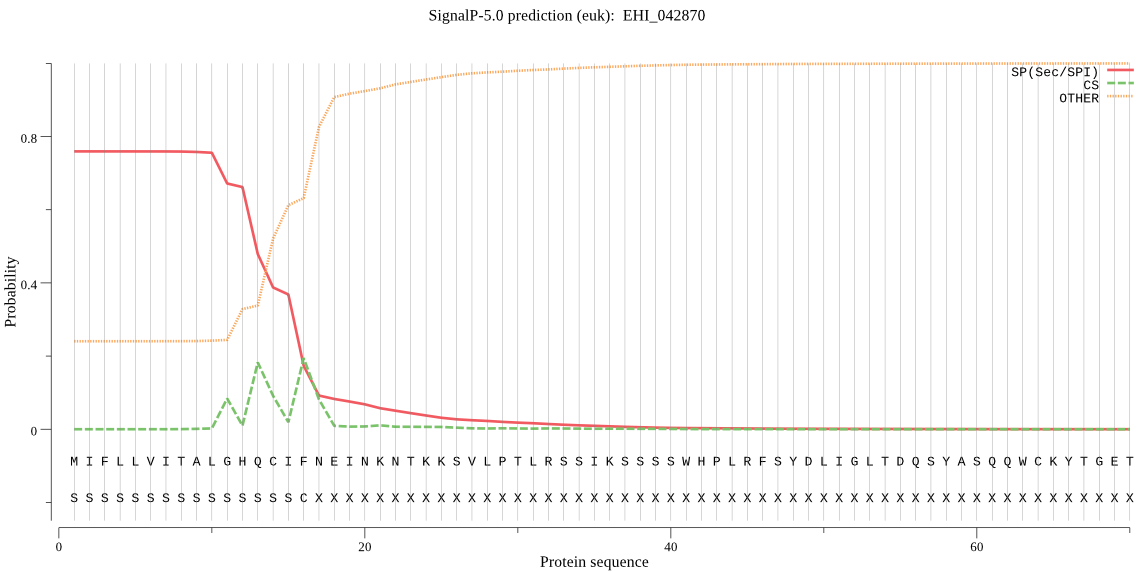
| EHI_042870 | SP | 0.063898 | 0.935254 | 0.000848 | CS pos: 14-15. HQC-IF. Pr: 0.2814 |
MIFLLVITALGHQCIFNEINKNTKKSVLPTLRSSIKSSSSWHPLRFSYDLIGLTDQSYAS QQWCKYTGETIILSSSNIYKCSEDDVLTKEKIEYIDRLFQSLNQSISEVFNVHTINYNKV IPGKKYCGEAVFPETYTIPEESDFHIMVTSHPHTSSGVFAYAAACYFGMDSQVNKRRSVM GYVNIVTSALEPTEERFRDQVGIIFHETMHALGFDGSLGVDKKVGDLNLKVVEDEIVVAR ARKHFGCDNMTYVPLEDGGGRGTANAHWERRIFISEIMNGITSNNPRVSEITLAYFEALG VYKPNYEMADALSWGRGLGCSFLEDCSKWPKQNGYYCRTGVRRCTNDRSAIGICDGNSFK ETLNKSYQHYGDPHIGGSDEIADNCIFVSPIETSYCYQKMSALSFDYYFTLEMLNHGQNF GDTSMCFESSLIKFIPFGINNYHCYSVACMNNSFYKISIDGRYYDCDKSIKVRGYGGKLI CADPKDICGNRVIETWPEIDSVENKTISVGDVLVINGRNLDVVQKVYIDYTSCKILHKSK EQLQIEVDYHDPFISFISFEASLMIEANESVNSCYDHLIKIKVPTYYIFLNFGDWVMTNY IFIAVIPVWIIFFILLFGYYTLKVVVIRQAEEVREAKREINLRLGNLKDKPQKKKQSFCV MF
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EHI_042870 | 178 S | NKRRSVMGY | 0.992 | unsp | EHI_042870 | 178 S | NKRRSVMGY | 0.992 | unsp | EHI_042870 | 178 S | NKRRSVMGY | 0.992 | unsp | EHI_042870 | 358 S | CDGNSFKET | 0.995 | unsp | EHI_042870 | 34 S | TLRSSIKSS | 0.997 | unsp | EHI_042870 | 40 S | KSSSSWHPL | 0.994 | unsp |