

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_050570 | SP | 0.010226 | 0.989439 | 0.000335 | CS pos: 20-21. SWA-AK. Pr: 0.8168 |
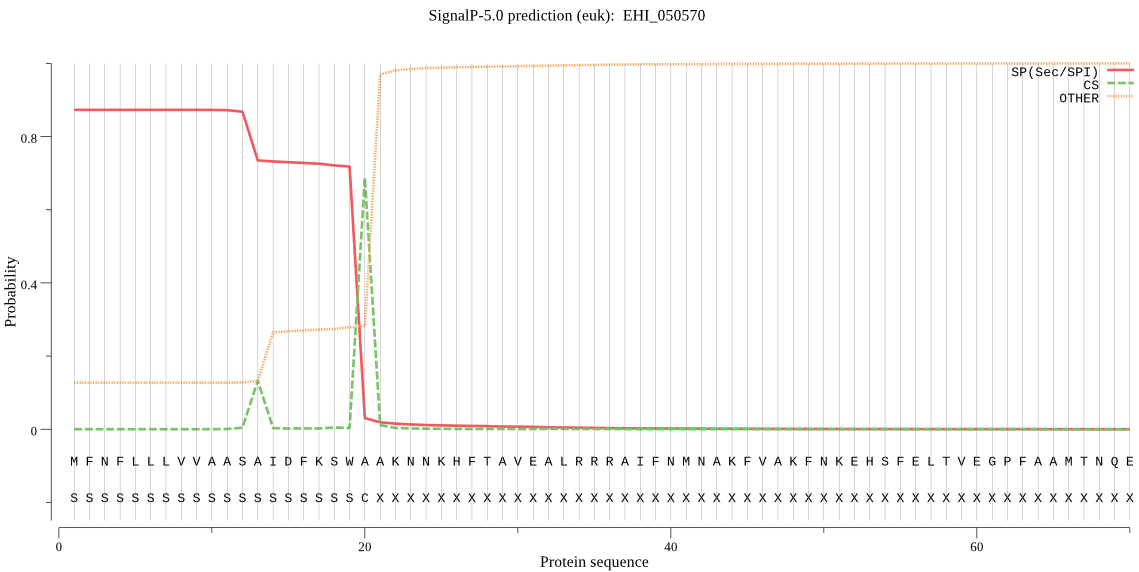
MFNFLLLVVAASAIDFKSWAAKNNKHFTAVEALRRRAIFNMNAKFVAKFNKEHSFELTVE GPFAAMTNQEYNNLLKTHETEAAADSVYDNTIITASSKDWRAEGKVTPVRDQGNCGSCYS FSSLAVLESRLLIAGSKYNQNNLDLSEQQIVDCSAANNGCNGGSLSATYLYVKNKGVTDE ASYPYTATKGTCKAFTPKVQTTGLTHVTPTEEALTAALAEGPVAVCIDAGKASFQLYKNG VYDEPKCSKTVNHGVAAVGYGSQDGQDYYIVKNSWGTSWGDKGYILMSRNKNNQCAIASV AYFPTGAHDAN
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EHI_050570 | T | 210 | 0.571 | 0.593 | EHI_050570 | T | 201 | 0.552 | 0.080 | EHI_050570 | T | 208 | 0.551 | 0.082 | EHI_050570 | T | 196 | 0.547 | 0.079 | EHI_050570 | T | 202 | 0.521 | 0.290 | EHI_050570 | T | 205 | 0.508 | 0.062 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EHI_050570 | T | 210 | 0.571 | 0.593 | EHI_050570 | T | 201 | 0.552 | 0.080 | EHI_050570 | T | 208 | 0.551 | 0.082 | EHI_050570 | T | 196 | 0.547 | 0.079 | EHI_050570 | T | 202 | 0.521 | 0.290 | EHI_050570 | T | 205 | 0.508 | 0.062 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| EHI_050570 | 86 S | AAADSVYDN | 0.995 | unsp | EHI_050570 | 96 S | IITASSKDW | 0.997 | unsp |