

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
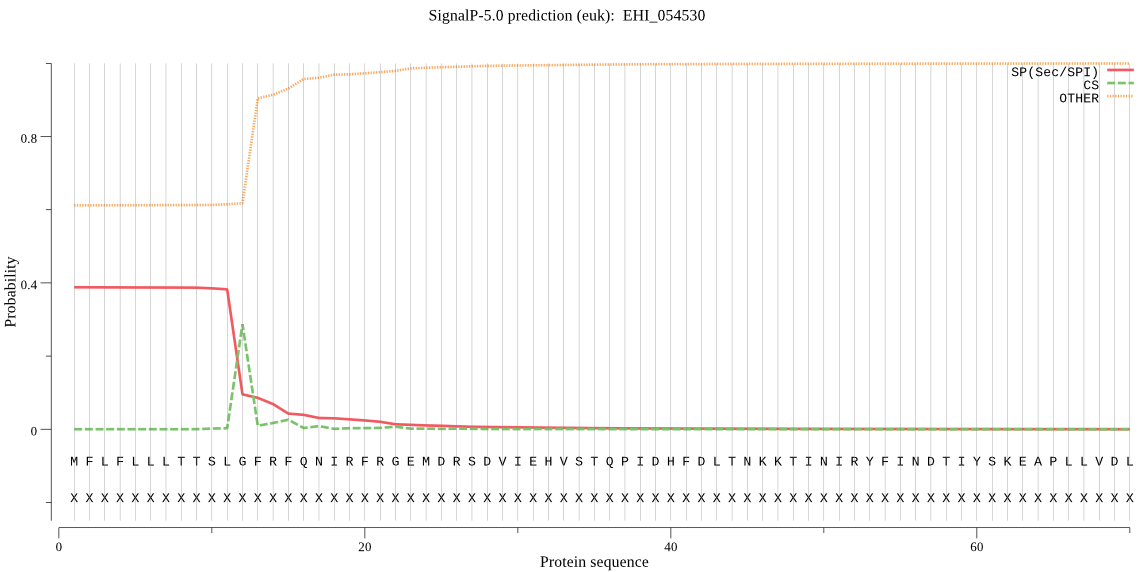
| EHI_054530 | SP | 0.273708 | 0.675459 | 0.050833 | CS pos: 12-13. SLG-FR. Pr: 0.4300 |
MFLFLLLTTSLGFRFQNIRFRGEMDRSDVIEHVSTQPIDHFDLTNKKTINIRYFINDTIY SKEAPLLVDLGGEGTQRAAAVGGRFVINKYAEKYNSLMLAIEHRFYGKSVPEGGLSQENL GYLSAAQALEDYIMIINQIKKEYQITGPVIVFGGSYSGNLATWIRQKYPNVVYAAVASSA PVYATSTFYEFLDVIYNDMGEKCGNAWKEATDSIEELFKTDSGKAQLKNDFKTCTEIKEE DDLTILIQQIQATMVNYPQYNGSYSLTIEGVCNILTTEGKTAYENMVDLMSHAFNEFGFE CAPSSYADMLTDMANTKTEEEGNRLASTRSWAWQICSEYSYFQPVNESLPFSKRLNNEFY YLLCKDIFNVDKQRLDRRVYHTNLMYGGYKPKATNVAYTSGSTDPWSPLAKHETLPSDIN CYASYIKGTAHCADLYAEKDTDPEQLKQQRMETAQFIDELISRYNN
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EHI_054530 | 213 S | EATDSIEEL | 0.996 | unsp |