

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_055660 | OTHER | 0.964206 | 0.000568 | 0.035226 |
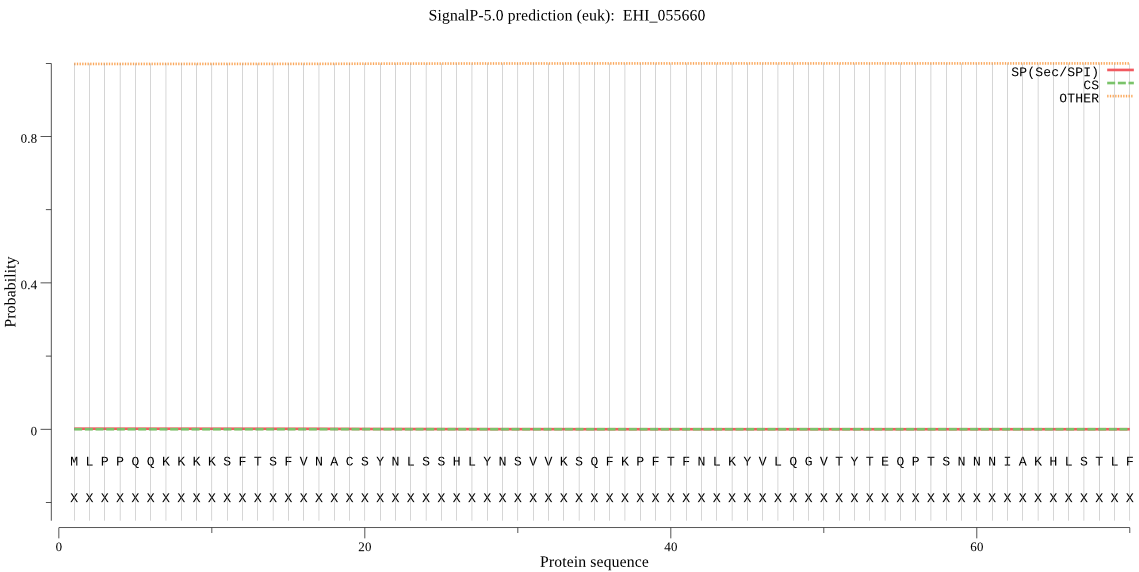
MLPPQQKKKKSFTSFVNACSYNLSSHLYNSVVKSQFKPFTFNLKYVLQGVTYTEQPTSNN NIAKHLSTLFRITYRNGFTYHLPHCSLTTDAGWGCTLRSIQMLFLNSLIRLQEPNPGFGD DAAEKVQQNFIIHSMEERREYVQLIEDTPKQEAVLSLYKMFNLKIVRQNNQKGTNYLSPS TCAIALSQLVEMWDQRPCHVIYSNTFPKEIQPNTLLMISAPLNEKTISCLDNTFVGGVVC GVDTKAIYVCGRTGNMLLLLDPHFVQKAHEDGDFDIVDYSVKPSDLRMVRLTELVFGNCI WGILVREDNIEILKNWCKTSLGVTNEEEIREMVSVGNEEGFEVLDF
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EHI_055660 | 280 S | IVDYSVKPS | 0.991 | unsp |