

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_062480 | SP | 0.031506 | 0.968428 | 0.000066 | CS pos: 18-19. CFG-DE. Pr: 0.9419 |
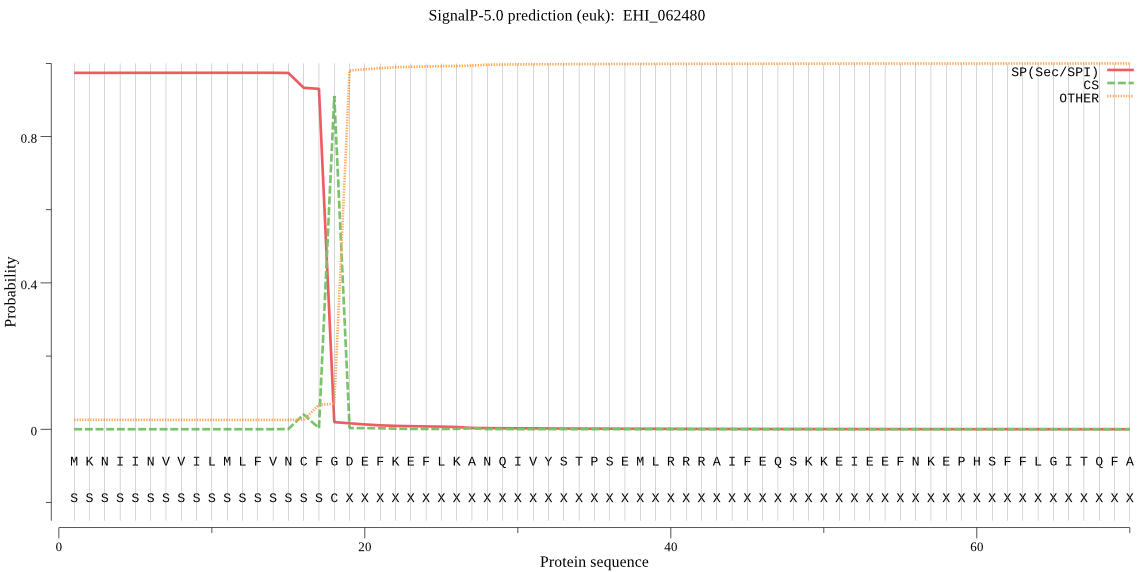
MKNIINVVILMLFVNCFGDEFKEFLKANQIVYSTPSEMLRRRAIFEQSKKEIEEFNKEPH SFFLGITQFADKTDEEFNQMFSMKMDRQDEFSMMSTDEEENKNESDDVYMEYNLSQDEET YGREMRRKVRGKKYRGVRFMRKVHVPKKYRIGRKWQFNKKKDIVKELPEGIDFRKFGKLT YIREQTGCGGCWSFASVCALESRYLIDYNLTVDDVGRTWALSEQQLLDCCIENNGCEGGS MERSFRCMNRTKGVMQRIRYPYEAETQDCKEFNNEYKEVTLGGYALVPRGNERALMSAIH KFGVLGIGLDTRSKLFKHYRGGIYYNEECTRRGLSHAMNLVGYGTTKEGQKYYIIRNSWG DWKWGEDGYMRLYRGGNHCGVATNAFFPLFVRRVNDDPKPMSHTETMIKDLINRHRLGWI
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EHI_062480 | 402 S | PKPMSHTET | 0.996 | unsp | EHI_062480 | 402 S | PKPMSHTET | 0.996 | unsp | EHI_062480 | 402 S | PKPMSHTET | 0.996 | unsp | EHI_062480 | 48 S | IFEQSKKEI | 0.993 | unsp | EHI_062480 | 358 S | IIRNSWGDW | 0.997 | unsp |