

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_064200 | OTHER | 0.999573 | 0.000383 | 0.000044 |
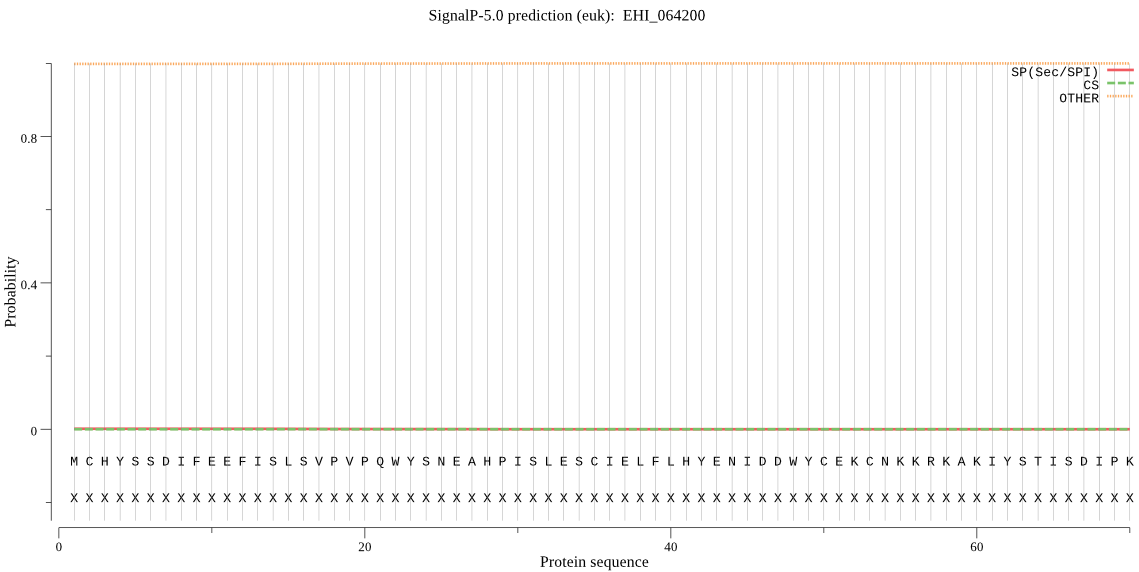
MCHYSSDIFEEFISLSVPVPQWYSNEAHPISLESCIELFLHYENIDDWYCEKCNKKRKAK IYSTISDIPKVLIIQLVRIYSKKYPIQQVLFPLNDLVVQTSPNHFDFYDLYSFITHTGSL SKGHYISWNKVSNQWYCCNDENVSQGNPTTSSDTVYILFYKRKVNSAGYNK