

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_068090 | SP | 0.060982 | 0.922140 | 0.016878 | CS pos: 18-19. SLN-QQ. Pr: 0.3880 |
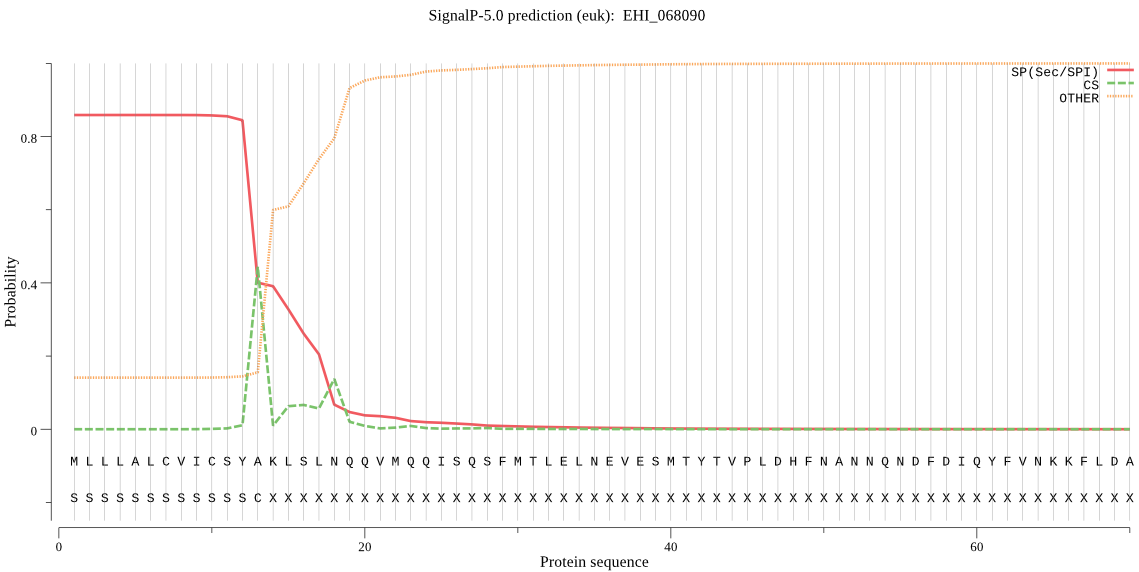
MLLLALCVICSYAKLSLNQQVMQQISQSFMTLELNEVESMTYTVPLDHFNANNQNDFDIQ YFVNKKFLDANDPNAPLFVLLGGEGPASPKVLQNNYVIDSLAKKHKGLMLSVEHRFYGAS TPSLEMDKLIYCTAEQALMDYVEVISHVQEENNLVGHPVIVLGGSYSGNLAAWMRQKYPN VVEGAWASSAPVEAVVDFYQYLEVVQNALPKNTADLLSFAFEQWDKMTTTEEGRKELGKI FNTCTEFGEKDIQTFAESIGTALSGYVQYNSSNWKPSYESTDSICAEINEDIVNKYPLFI KEKYNPEWADKECTPSSQEESYKTLQNTSTYAEGNEDASGRSWFFQTCIAYGYYQAVSEQ SSVKWGKLNQLQGSIDMCKDIYGIDKDTLYNAVDHINVRYGGKKPCVTNVAFTNGNTDPW HALGVTESDHQEGNLVQLIDRTSHCSDLYSEKENDVPELKKARHNELKFFAQVLANVPQN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EHI_068090 | 317 S | CTPSSQEES | 0.996 | unsp | EHI_068090 | 317 S | CTPSSQEES | 0.996 | unsp | EHI_068090 | 317 S | CTPSSQEES | 0.996 | unsp | EHI_068090 | 362 S | SEQSSVKWG | 0.995 | unsp | EHI_068090 | 450 S | SDLYSEKEN | 0.995 | unsp | EHI_068090 | 88 S | EGPASPKVL | 0.99 | unsp | EHI_068090 | 277 S | NWKPSYEST | 0.996 | unsp |