

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
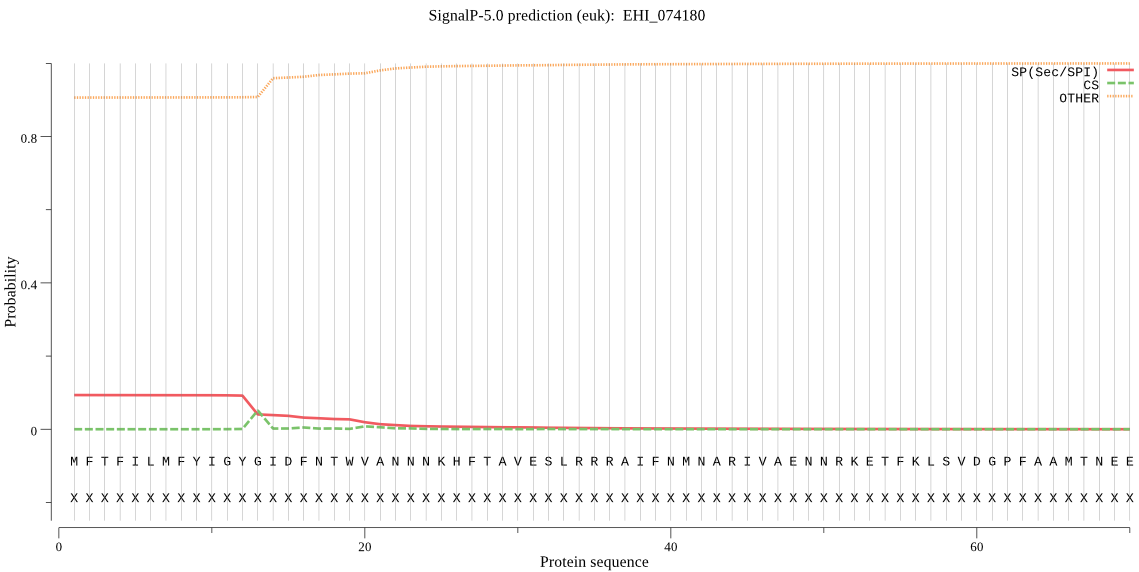
| EHI_074180 | SP | 0.278038 | 0.720806 | 0.001156 | CS pos: 13-14. GYG-ID. Pr: 0.4600 |
MFTFILMFYIGYGIDFNTWVANNNKHFTAVESLRRRAIFNMNARIVAENNRKETFKLSVD GPFAAMTNEEYNSLLKLKRSGEEKGEVRYLNIQAPKAVDWRKKGKVTPIRDQGNCGSCYT FGSIAALEGRLLIEKGGDSETLDLSEEHMVQCTREDGNNGCNGGLGSNVYNYIMENGIAK ESDYPYTGSDSTCRSDVKAFAKIKSYNRVARNNEVELKAAISQGLVDVSIDASSVQFQLY KSGAYTDKQCKNNYFALNHEVCAVGYGVVDGKECWIVRNSWGTGWGEKGYINMVIEGNTC GVATDPLYPTGVEYL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EHI_074180 | 80 S | KLKRSGEEK | 0.991 | unsp |