

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
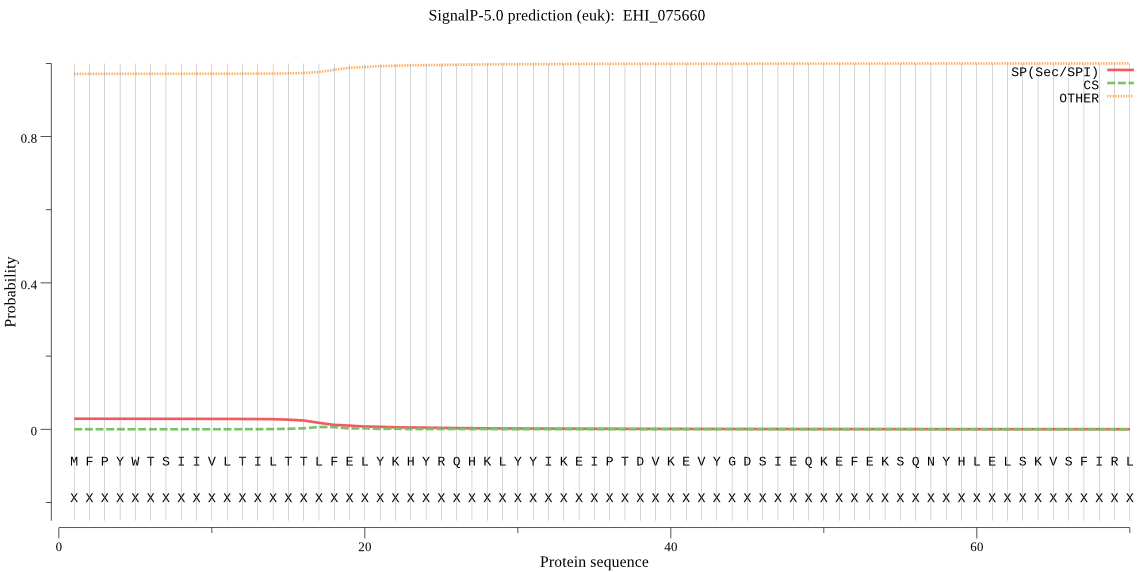
| EHI_075660 | OTHER | 0.996134 | 0.003455 | 0.000412 |
MFPYWTSIIVLTILTTLFELYKHYRQHKLYYIKEIPTDVKEVYGDSIEQKEFEKSQNYHL ELSKVSFIRLTISFIINMYVLCSPILRIIWDFSTIYNQFLTSIIFIIIFDFISTIISIPF KLYTTFIIREKYGMNNMSLIVFIKDFIKSFILETILNLIIITLLYFVSETQNLALYLWFG IMTLNIIISLIFVPFVIPLFYKKTPLQEDQYKNEIESKLNEVNFPLKSVSVIDASSKAKE GNAFFSGLFGKRDLVLFDTLITTCSSDELVDIVLHEVGHCKHYHIFKLLGIQSIQFFIIF KFIEFFLLDEALYTQFGFDQKVVVVGFILLQSLLEPFMEIVSLGINFISRNFEYQADVYA TKHGNHQLASALIKLQKNNLSAYVVDPFVSTIENSHPNLVERIQAINKIIKDMKSD
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EHI_075660 | 230 S | LKSVSVIDA | 0.996 | unsp |