

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_084060 | SP | 0.021199 | 0.978795 | 0.000007 | CS pos: 15-16. SEG-NS. Pr: 0.6331 |
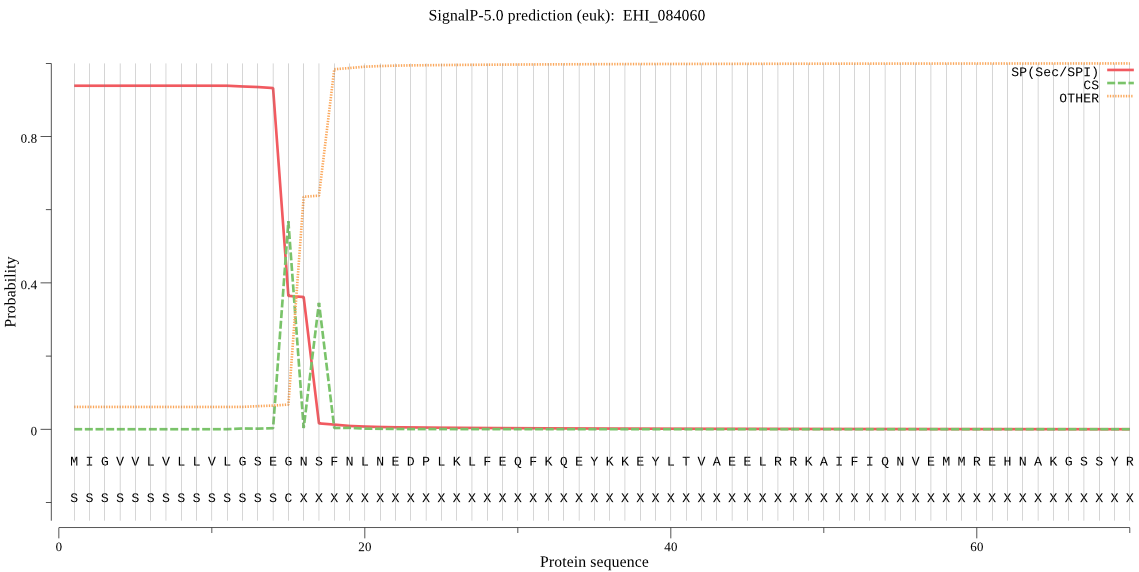
MIGVVLVLLVLGSEGNSFNLNEDPLKLFEQFKQEYKKEYLTVAEELRRKAIFIQNVEMMR EHNAKGSSYRMGINKFADMESNELLATSSIIQSNRRMGAKRIQEMIRMEKVYSTKQTLKQ AVPDNYTLCTSEAEYNYCGTNNIDQNLCGGCYAIATAHHLSTIYAYLTKQVDNNKPKRKM LSAQQLLDCNTESWGGCGGGFAEDVLDSVDGIYYADDYPFIDADKDCENVTDCAKYRHEC KSLEIDYPLKFATSNKFTHFANKNWEEIKEIIYKYHGFISAMKVTDSLLRYVGGIYKSES CTGKITDHIVVVDGYGECDGHKFLWVRSSWGNDWGVENGHFKIDWNTLCGINGVDEYDDG SQLQNNLYVEMELGTGDTAEYNPKDGAGDDNKDPIGCVIVSPSGDPSNSPEDNKTTGVVM ALFVFLMIFFI
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| EHI_084060 | 113 S | EKVYSTKQT | 0.992 | unsp | EHI_084060 | 409 S | DPSNSPEDN | 0.998 | unsp |